Difference between revisions of "Part:BBa K3332047"
Huangzinuo (Talk | contribs) |
AnnaTaylor (Talk | contribs) |
||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3332047 short</partinfo> | <partinfo>BBa_K3332047 short</partinfo> | ||
| − | The iNAP is fused at N-terminal with INPNC anchoring protein. We use K880005 to construct the expression system and anchor iNAP on the surface of E.coli. | + | The iNAP is fused at N-terminal with INPNC anchoring protein. We use K880005 to construct the expression system and anchor iNAP on the surface of ''E.coli''. |
===Biology=== | ===Biology=== | ||
Revision as of 23:26, 27 October 2020
J23100-RBS-INPNC-iNAP-terminator
The iNAP is fused at N-terminal with INPNC anchoring protein. We use K880005 to construct the expression system and anchor iNAP on the surface of E.coli.
Biology
GRHPR, a glyoxylate reductase from human liver, can reduce glyoxylic acid when NADPH is used as cofactor. GRHPR converts glyoxylic acid while consuming NADPH. NADPH is a suitable target compound that can be detected by the signal of fluorescence or OD340.[1]
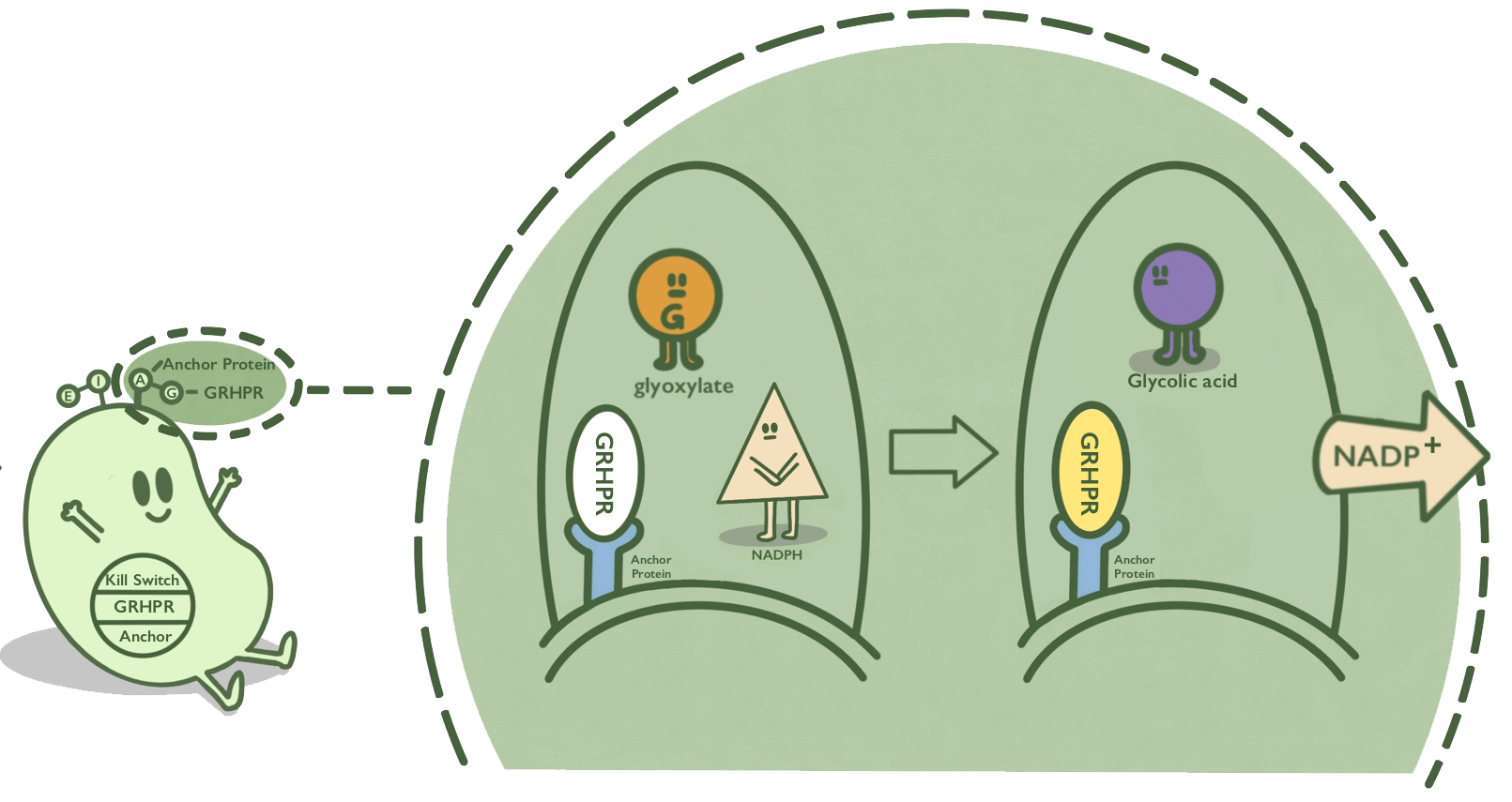
- Fig 1. GRHPR mechanism.
Usage
By codon optimization and adding a 6His-tag, the sequence suitable for expression in E. coli was constructed, and we hoped that it could reduce glyoxylic acid in E. coli to get fluorescence signal in the next processes we design.
The coding sequence of target gene was inserted into an expression vectors with BBa_K880005(BBa_J23100 & BBa_B0034) to obtain BBa_K3332056. We transformed the constructed plasmid into E. coli BL21 (DE3) to verify its successful heterologous expression.
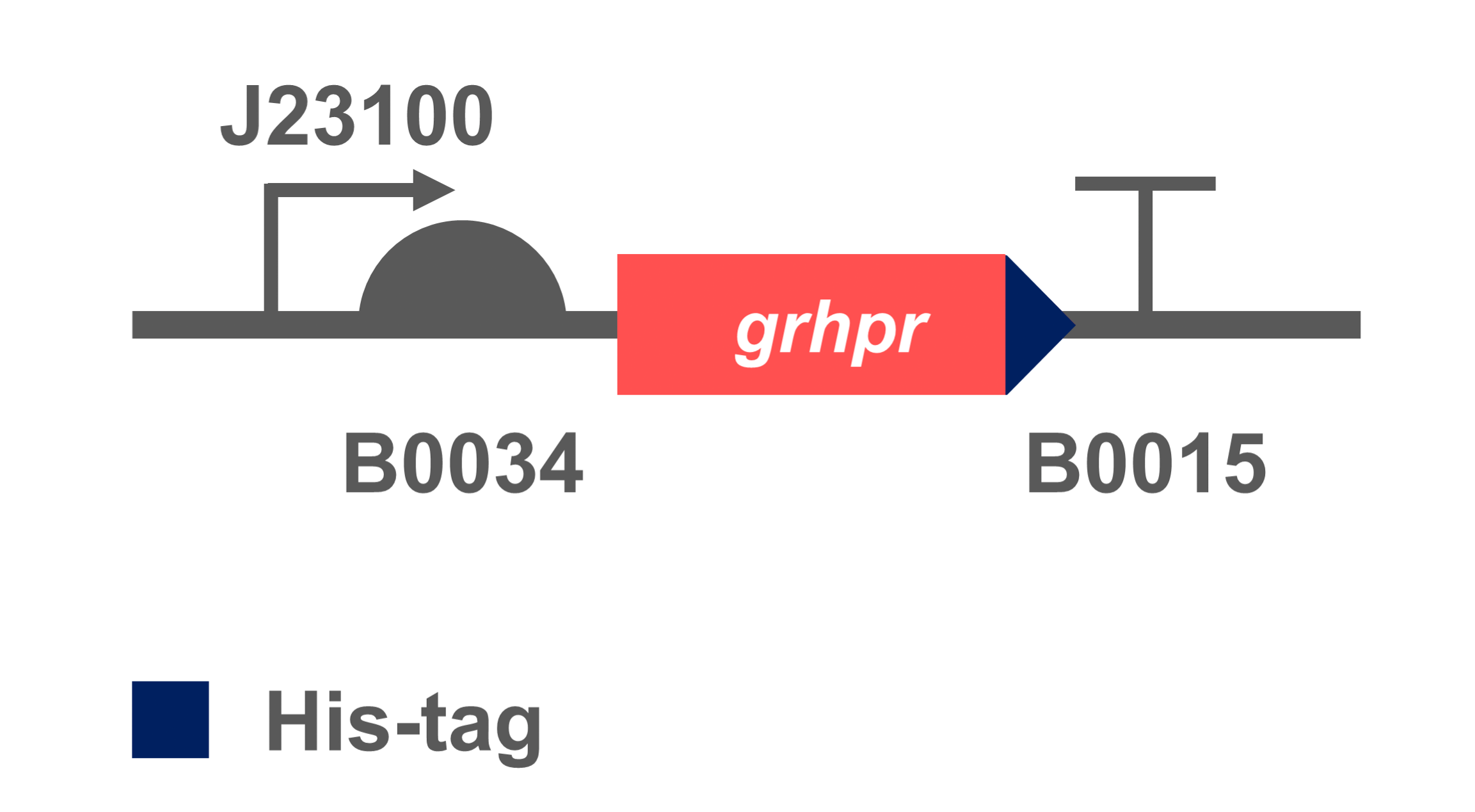
- Fig 2. Gene circuit of GRHPR.
Characterization
1. Identification
After receiving the synthesized DNA, restriction digestion was done to certify that the plasmid was correct, and the experimental results were shown in figure 3.
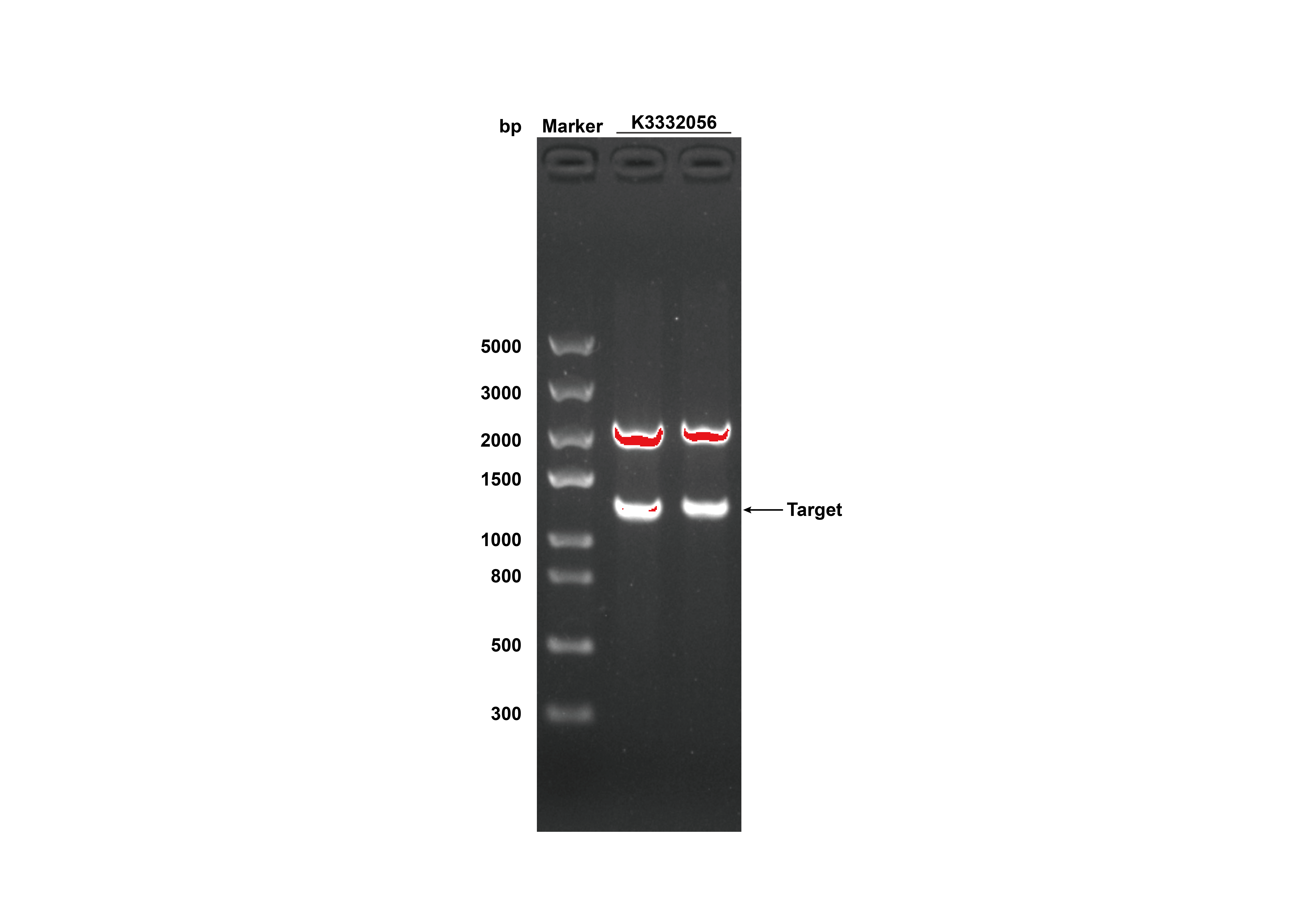
- Fig 3. DNA gel electrophoresis of restriction digest products of GRHPR-His-pSB1C3 (Xbal I & Pst I sites)
2. Purification and Proof of the expression
We used J23100 promoter to highly express GRHPR-Histag in E. coli in our composite part BBa_K3332056. Then, we used GE AKTA Prime Plus FPLC System to get purified GRHPR protein. We found an apparent protein peak in AKTA FPLC System and correct purified protein.
Then, our target bands are observed through SDS-PAGE and the result is shown in figure4.
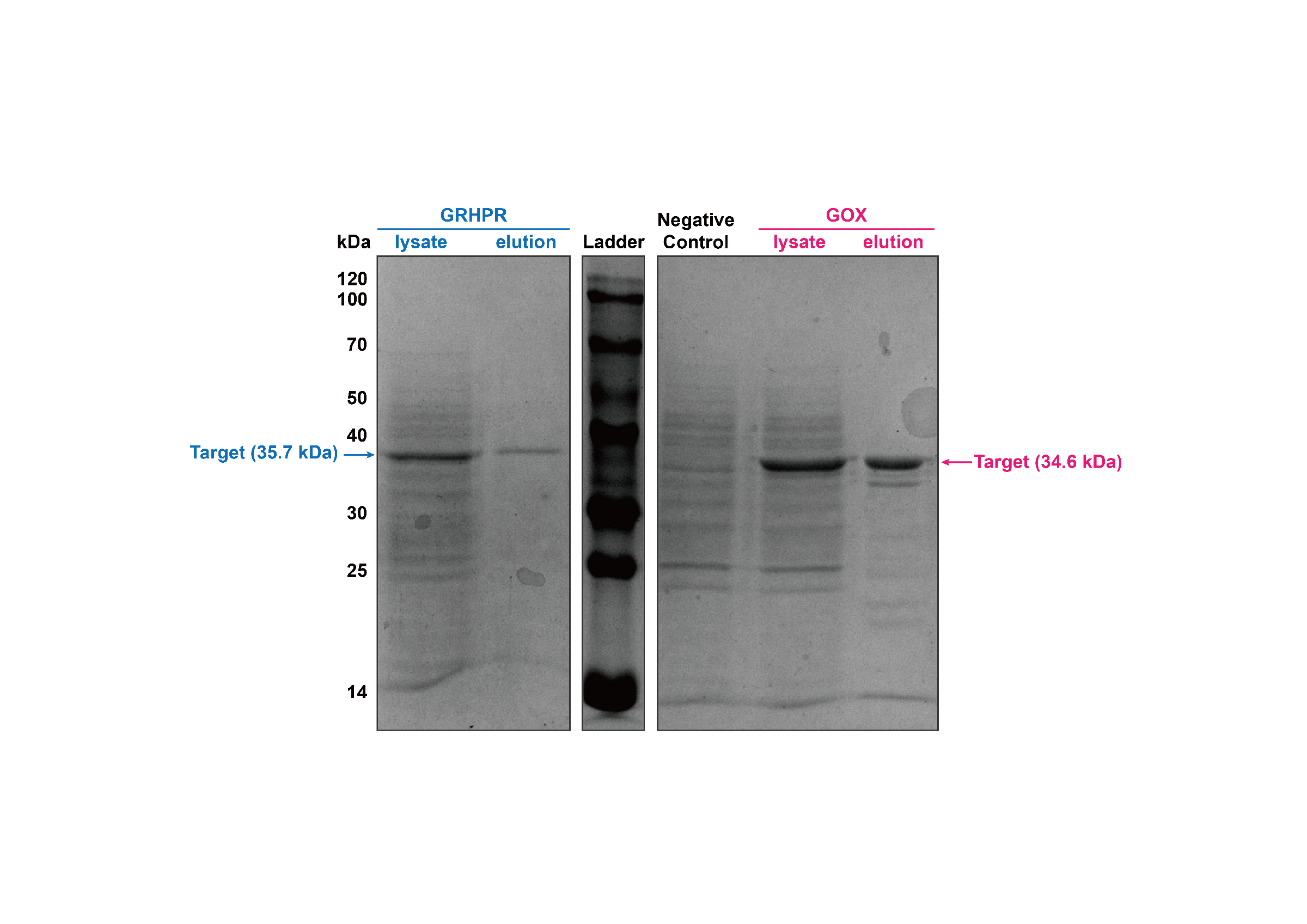
- Fig 4. SDS-PAGE of purification products of GRHPR-Histag-pSB1C3
3. Ability of consuming NADPH
We mixed glyoxylic acid solution, NADPH solution and purified GRHPR protein dissolved in Tris-HCl(pH=7.5). Then, we immediately measured OD340 changes of our samples. And when NADPH is consumed, OD340 declines.
The experimental result is shown on Figure 5. We can see the OD340 of samples adding GRHPR decrease very quickly while the OD340 of control stay almost the same.
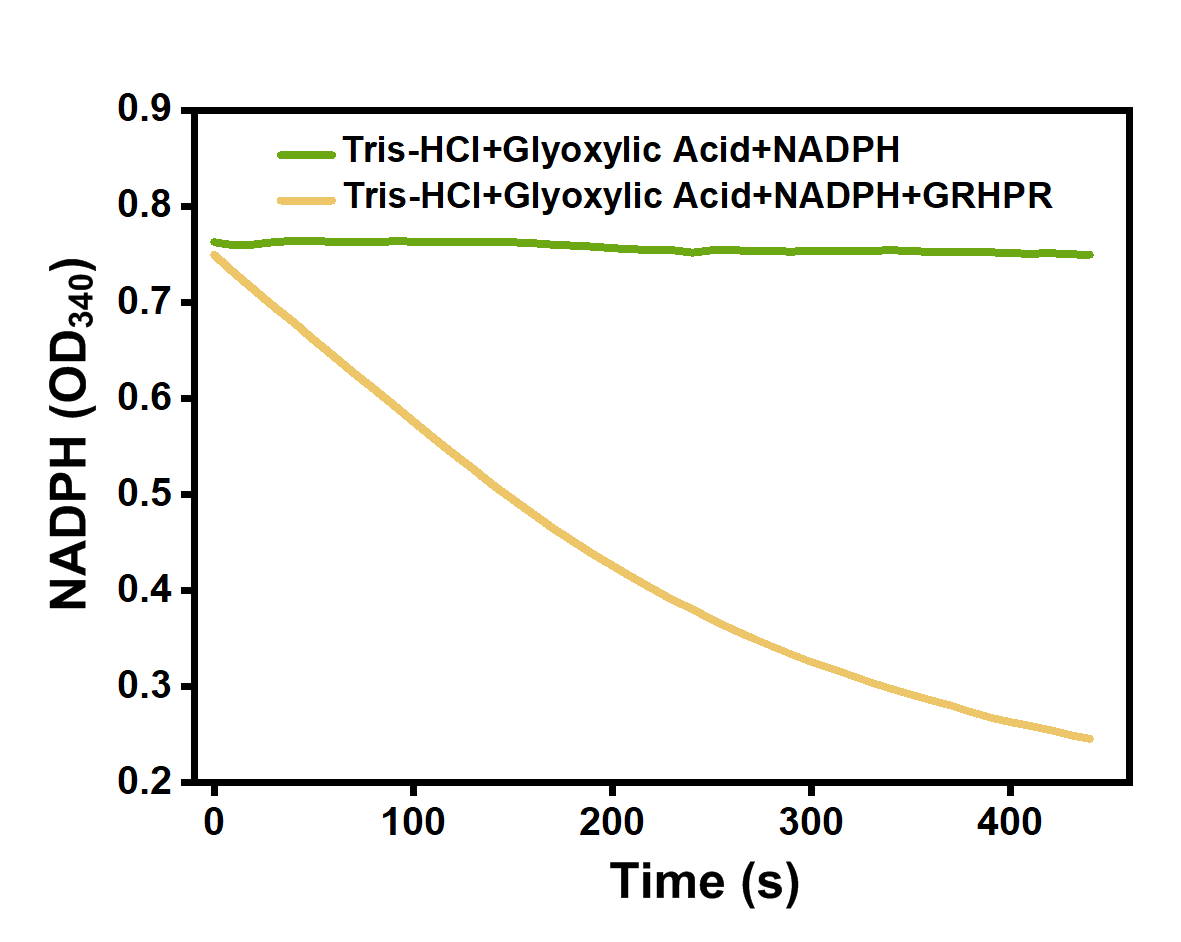
- Fig 5.' Enzyme activity of GRHPR.
4. Kinetic parameter determination
We successfully got OD340-Time curves of GRHPR in the presence of NADPH concentration and OD340-Time curves of GRHPR in the presence of glyoxylic acid concentration. Then we calculated relevant enzyme activity and drew 1/V-1/[NADPH] and 1/V-1/[glyoxylic acid] curves, from which we can obtain relevant Km and Vmax.
The result is shown in figure 6 and figure 7.
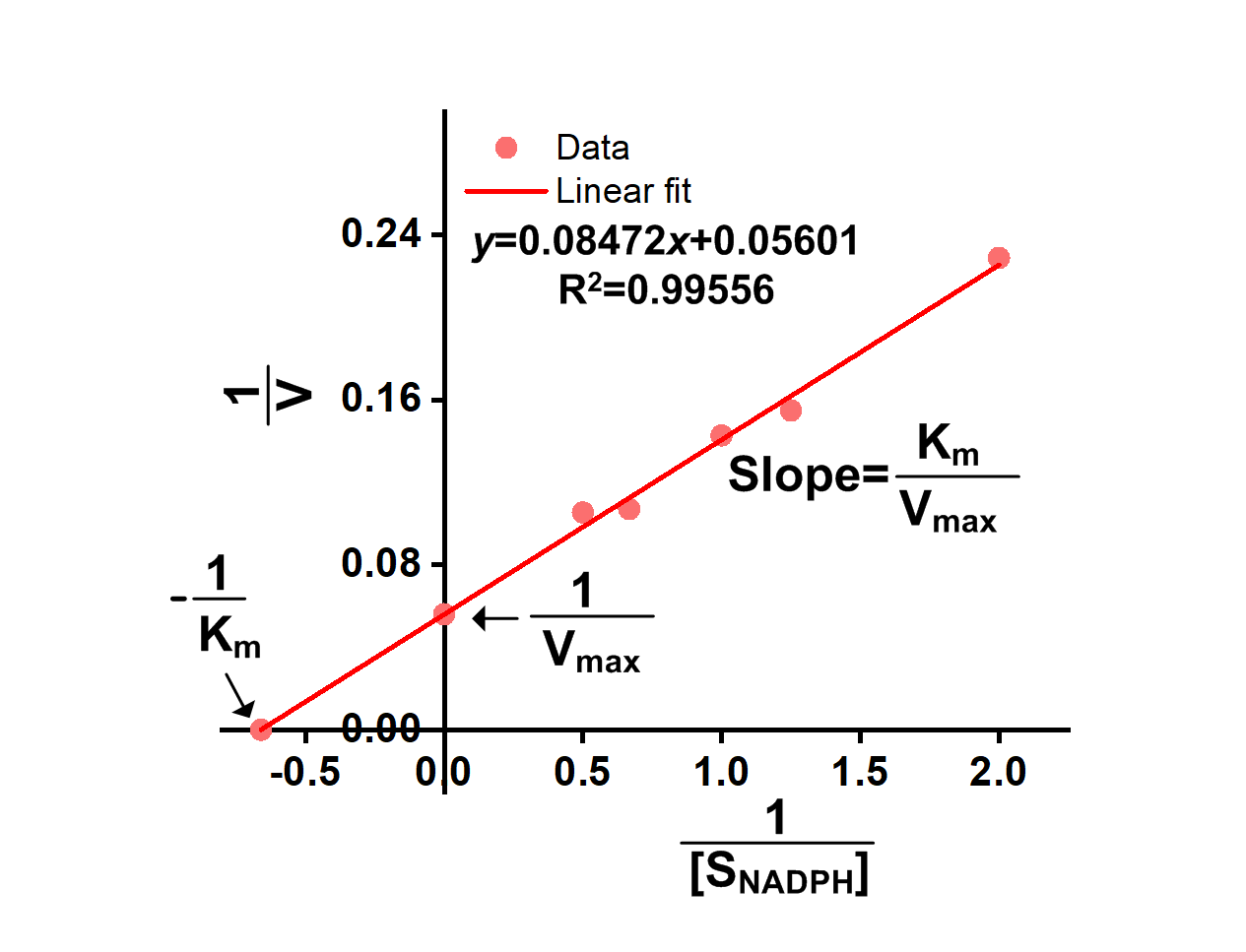
- Fig 6. 1/V-1/[NADPH] curve of purified GRHPR reacting with NADPH and glyoxylic acid
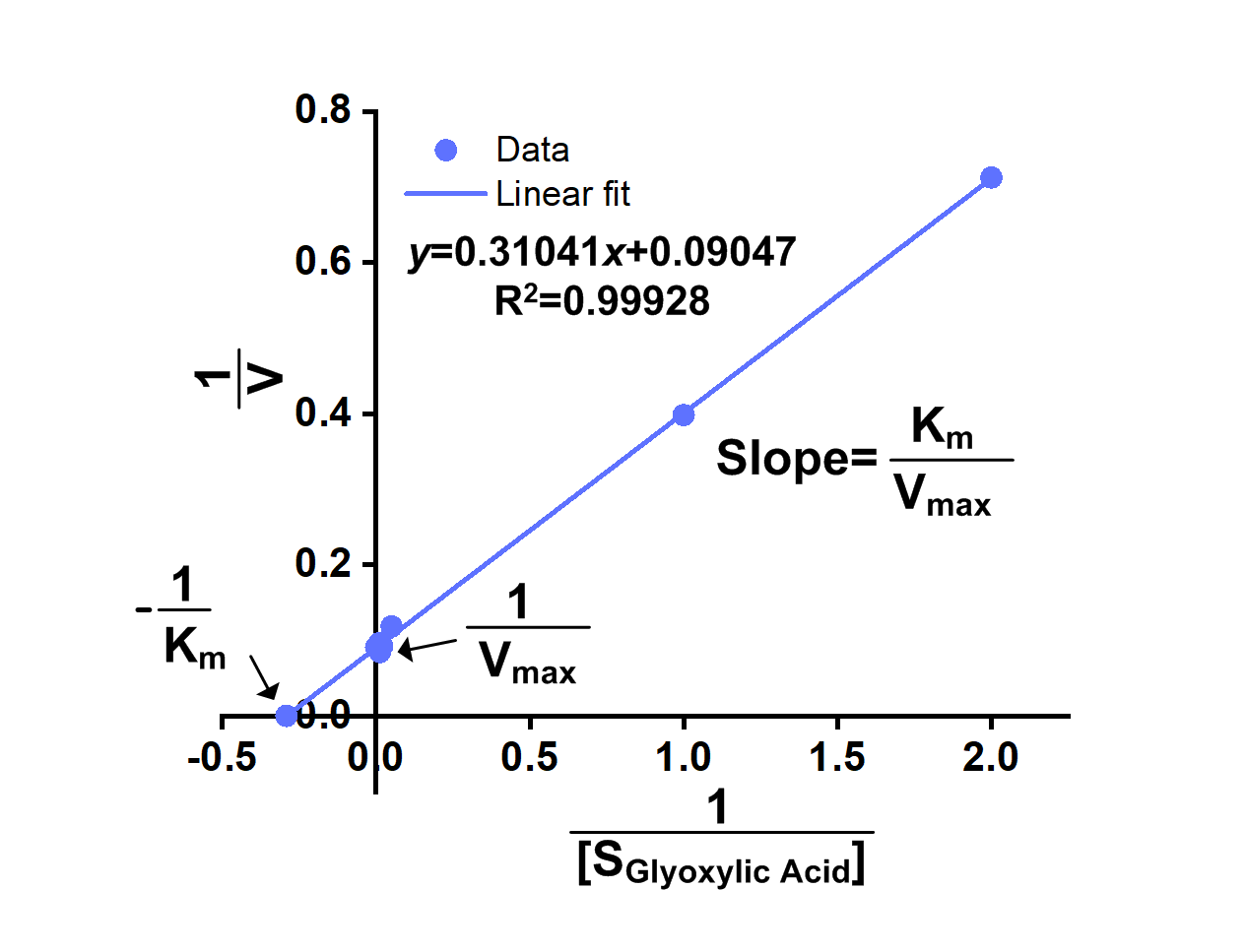
- Fig 7. 1/V-1/[glyoxylic acid] curve of purified GRHPR reacting with NADPH and glyoxylic acid
References
- ↑ Rumsby G, Cregeen D P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1999, 1446(3): 383-388.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 62
Illegal BamHI site found at 1314
Illegal XhoI site found at 1340 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 472
Illegal NgoMIV site found at 1080 - 1000COMPATIBLE WITH RFC[1000]
