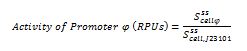Difference between revisions of "Part:BBa J23102:Experience"
| Line 156: | Line 156: | ||
|} | |} | ||
---- | ---- | ||
| + | |||
| + | |||
| + | {|width='100%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_J23100 AddReview 5</partinfo> | ||
| + | <I>University of Texas at Austin iGEM 2019</I> | ||
| + | |width='60%' valign='top'| | ||
| + | ====Characterization of the Anderson series through burden monitoring by the University of Texas at Austin's 2019 iGEM team==== | ||
| + | |||
| + | |||
| + | ===Description=== | ||
| + | Our team transformed the Anderson series of RFP reporters (J23101, J23113, J23104, J23107, J23117 in pSBC13 backbone) into our constitutive GFP burden monitor <I>E. coli</I> strain and measured the relative burden of each part. They contained a constitutive GFP sequence in the genome which serves as a way to measure the constructs’ ribosome allocation. Our results show a certain reduction ingrowth rate for each part as a result of ribosome misallocation away from the genome and towards the plasmid containing the construct. The promoter strengths associated with each RFP reporter construct shows that parts with stronger promoters express less GFP and have a reduced growth rate when compared to the constructs containing weaker promoters. The promoters associated with these RFP reporters were used to create a series of BFP reporters, also transformed into our GFP burden monitor strain, and create the regression on the figure below. For more information on characterization of these parts through burden monitoring and evolutionary stability experiments, visit our team’s wiki page: [https://https://2019.igem.org/Team:Austin_UTexas] | ||
| + | |||
| + | [[Image: AndersonCharacterization.jpg|450px]] | ||
Revision as of 04:09, 10 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J23102
Unexpected LuxR-AHL repressible behaviour
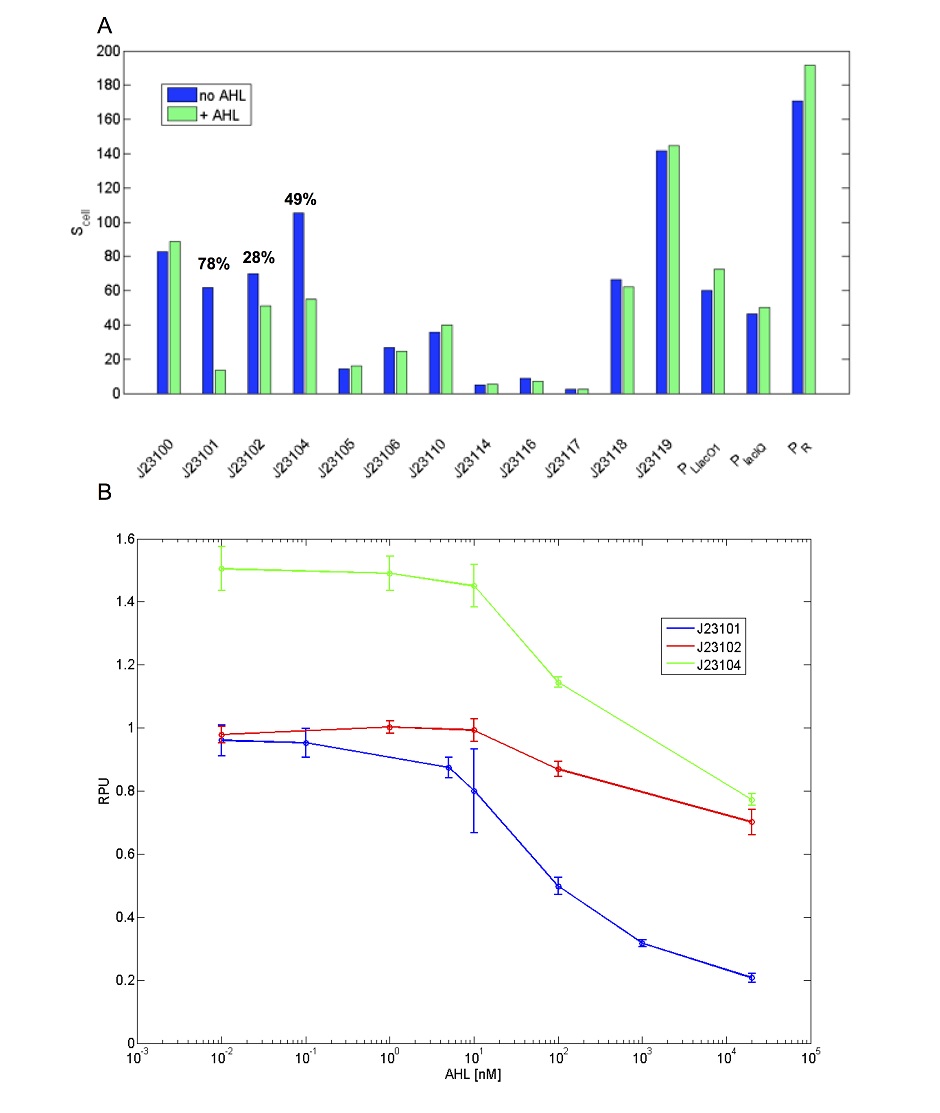
This promoter shows activity repression as a function of AHL in presence of LuxR [ADD REFERENCE LINK]. Results are shown below for a set of widely used promoters tested in the same conditions (TOP10, M9 medium supplemented with casamino acids and glycerol, assayed in a microplate reader). Promoters drive the BBa_I13507 RFP expression device. The TACTAGTG scar is present between promoter and RBS, except for PLlacO1 (BBa_R0011), PlacIQ (BBa_I14032) and PR (BBa_R0051) where the scar is TACTAGAG. RFP was measured and used to compute Scell (arbitrary units) or RPU values. Promoters were assembled in pSB4C5 and were co-transformed with BBa_S03119 in pSB3K3 in TOP10. Blue bars represent the activity when no AHL was added to the medium, while green bars represent the repressed activity (AHL 20 µM was added to the medium). Only BBa_J23101, BBa_J23102 and BBa_J23104 showed repression and, for them, percent repression is reported.
Evaluation of Anderson promoter J23102 in B. subtilis by iGEM-Team LMU-Munich 2012
This Anderson promoter was evaluated without fused RFP with the lux operon as a reporter in B. subtilis. See the new BioBrick BBa_K823006 without RFP and have a look at the [http://2012.igem.org/Team:LMU-Munich/Data/Anderson Data] from the evaluation in B. subtilis.
User Reviews
UNIQ10d1c4d3e0411157-partinfo-00000000-QINU UNIQ10d1c4d3e0411157-partinfo-00000001-QINU
iGEM WHU-China 2013 construction of tandem double promoters

figure 1. The seven different combinations of double tandem promoters,from left to right, are J23106,J23102-J23102,J23102-J23106,J23106-J23102,J23106-J23106,J23106-J23116,J23116-J23102 and J23116-J23106.
We use consecutive promoter BBa_J23102 as a basic part to construct tandem double promoters BBa_k1081002(J23102-J23102),BBa_k1081003(J23102-J23106),BBa_k1081004(J23106-J23102) and BBa_k1081007(J23116-J23102).
iGEM CINVESTAV_IPN_UNAM CHARACTERIZATION OF IGEM DISTRIBUTION BIOPARTS
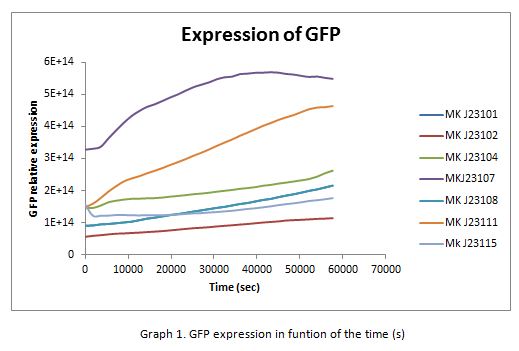
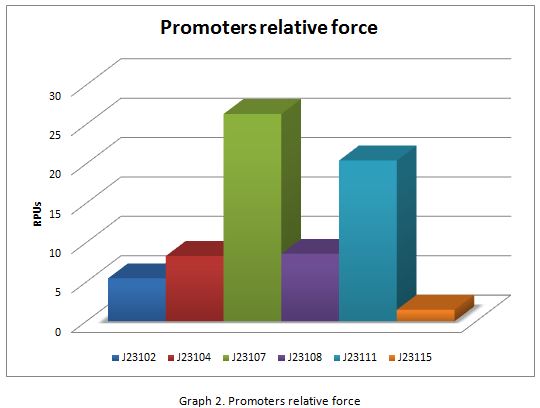
For contribute to the parts registry our team decided to make the characterization of constitutive promoters belonging to the family isolated from a small combinatorial library (J23101 , J23102, J23104, J23107, J23108, J2311, and J23115) which were attached to GFP to determine promoter activity, using the equipment Victor X3 Multilabel Plate Reader.
Fig. 1 Construction of the promoter J23102 expressing GFP.
Methods
With the selected colonies, an overnight culture was made in M9 media(minimal media supplemented with 0.2% CAA). After 12 hours the culture was transferred to a 96 well plate at a 1:10 dilution (20 μl of culture and 180 μL of fresh M9 medium). OD and fluorescence measurements of the selected colonies were performed at intervals of 30 minutes for 16 h. From the results the PopS were calculated (polymerases per second).
Modeling
The ecuations used for calulated de promoter activity were based on (R. K. Jason et. al 2009).
Results
In the following graphs there is shown the GFP expression in function of th time and the realtive promotor intensity.
With the previous results of the characterization of the promoters there is concluded that the promoter J23107, is the strongest because it produces more RPUs”
iGEM Bulgaria 2017 Regulation of the J23102 promoter activity by CRISPi
We offer an improvement to the J23102 part. We designed 2 gRNAs that target this promoter (TTGACAGCTAGCTCAGTCCT and TAGTAGCTAGCACAGTACCT) and cloned them into our gRNA expression vector (Part:BBa_K2515002). These constructs gave us the ability to control the J23102 promoter activity in a strain with dCas9 unther teh control of an inducible arabinose promoter. We believe that this type of regulation can be transffered to the other members of this promoter family if one redesign the gRNAs.
Determination of Noise Levels in Constitutive Promoter Family Members
(Characterized by SDU-Denmark)
Fluorescence microscopy and flow cytometry revealed decrease in fluorescence over time for members of the constitutive promoter family.
The expression levels and the noise of four different members of the Anderson promoter collection and their RFP reporter systems, were studied by fluorescence microscopy. These were, in increasing promoter strength, BBa_J23114, BBa_J23110, BBa_J23106, and BBa_J23102.
Additionally, the change in RFP expression levels and noise during growth were tested for the promoters with the highest and lowest relative promoter strength by flow cytometry and qualitative analysis by fluorescence microscopy. Combining these two techniques, the expression and noise levels for the promoters were determined as follows:
- The weak promoter, BBa_J23114, exhibited a relatively low expression of RFP, indicating low gene expression and an increasing high level of noise throughout growth.
- Both medium strength promoters, BBa_J23110 and BBa_J23106, displayed a moderate level of both noise and protein expression of the RFP reporter.
- The strong promoter, BBa_J23102, exhibited a comparatively high expression of the reporter RFP and an increasing high level of noise throughout growth.
For these experiments, the promoters controlling an RFP reporter system were cloned into E. coli strain MG1655 on pSB1A2. The cultures were grown in LB medium containing 50 µg/mL ampicillin and examined by fluorescence microscopy, using an Olympus IX83 with a photometrics prime camera and 200 ms exposure time, when cultures were at OD600=0.3-0.5. For flow cytometry, both LB medium with 50 µg/mL ampicillin and M9 medium with 100 µg/mL ampicillin were used. Excitation of RFP was at 561 nm, and emission was measured around 580 nm.
The images obtained by fluorescence microscopy, which are given in Figure 1, revealed that the weak promoter, BBa_J23114, presented high variability in the expression of RFP between cells, indicating a high level of noise. Contrarily, RFP expression was more uniform in the bacterial cells containing the medium promoter Bba_J23106. Similarly, another medium strength promoter BBa_J23110, showed uniform level of RFP expression, although less consistent than the observed for BBa_J23106. Expectedly, the strong promoter, BBa_J23102 exhibited high RFP expression in cells and a low level of noise was observed.

Figure 1.Fluorescence microscopy of RFP under the regulation of four members of the Anderson promoter collection, BBa_J23114, BBa_J23110, BBa_J23106 and BBa_J23102) on pSB1A2 in E. coli MG1655 at OD600=0.3-0.5. BBa_J23114 displayed a low and noisy expression of RFP, whereas both BBa_J23106 and BBa_J23110 showed a more uniform expression of medium intensity. The strongest promoter, BBa_J23102, exhibited a high level of consistent RFP expression.
The noise levels observed by fluorescence microscopy led to the design of an experiment, by which the cell population could be quantitatively studied. The change in expression levels and noise during growth of cell populations were tested for the weak and strong constitutive promoters, for which flow cytometry was used to assess the expression of the RFP reporter system. First, the cell populations were studied in LB media with 50 µg/mL ampicillin, thereby maintaining similar conditions as for the fluorescence microscopy experiment. All cultures were prepared from overnight cultures, obtaining a starting OD600=0.005.

Figure 2. Flow cytometric fluorescence measurements in arbitrary units as a function of time. Left: Cultures were grown in LB medium. Right: Cultures were grown in M9 minimal medium supplemented with 0.2% glycerol. Fluorescence of RFP expressed by the weak and strong constitutive promoters were measured relative to the negative control WT E. coli MG1655. Samples where measured in technical replicates and standard error of mean is shown, but are in several cases indistinguishable from the graph.
The data obtained in this experiment revealed that the strong promoter displayed a higher basal level of RFP expression than the weak promoter. Moreover, both promoters exhibited similar decrease in fluorescence over time, as seen in Figure 2, indicating that a considerable portion of the cell populations for both promoters lost their ability to fluoresce during growth, with one population containing around 60% non-fluorescent bacteria, clearly divided from the fluorescent portion. As expected, the strong promoter displayed a high level of RFP expression, roughly 500-fold of the non-fluorescent MG1655 control at 1 hour. A 10-fold decrease in fluorescence was observed from 1 hour to 4 hours. The RFP expression mediated by the weak promoter exhibited a similar decrease in fluorescence over time, though with measured fluorescence levels 5 times lower than for the strong promoter.
Comparing these findings to the fluorescence microscopy data, it was hypothesised, that the increasing noise could be caused by bacterial plasmid loss. This could be ascribed to degradation of ampicillin in the medium, since the mechanism behind ampicillin resistance relies on the β-lactamase mediated cleavage of the antibiotic, thereby relieving the selective pressure. Consequently, the bacteria, which have lost their plasmid could be able to compete with the bacteria still containing their plasmid.
The fluorescence microscopy for the strong promoter did not exhibit a considerable level of noise, however, this does not reject the hypothesis. The increase in noise was revealed by flow cytometry, as measurements were carried out at several times throughout growth. However, the fluorescence microscopy measurements were only carried out once in the exponential phase, where the noise was not prevalent yet.
To substantiate the hypothesis, the experiment was performed anew using M9 minimal medium supplemented with 0.2 % glycerol and 100 µg/mL ampicillin, in an attempt to decrease the plasmid loss rate and optimise the selection.
The resulting data revealed, that the decrease in fluorescence levels had a lower rate for both promoters when the bacteria were cultured in M9 minimal medium, seen in the right graph in Figure 2, than in LB medium, seen in the left graph in Figure 2. Furthermore, when the bacteria were grown in M9 minimal medium with double amount of antibiotics compared to the LB medium, they displayed a higher level of fluorescence. Some of these observations could potentially be ascribed to the increased selection, that was strived after in this experiment. Moreover, the difference between the measured fluorescence levels for the two reporter systems was notably lower. This could indicate, that the reason behind the difference in expression levels between the weak and strong constitutive promoters, is due to the fact that bacteria cloned with the weak constitutive promoter are more prone to plasmid loss. However, it was observed that the bacteria expressing RFP under control of the weak promoter, did in fact fluoresce with a lower intensity.
To verify the hypothesis regarding the plasmid loss, the cultures were plated on 50 µg/mL ampicillin LA selection plates as well as LA plates without antibiotic, 12 hours after incubation start. For comparison, cultures containing either chloramphenicol and kanamycin resistance cassettes, were likewise plated out after similar treatment. The average plasmid loss measured as non-resistant CFU as a percentage of the total population is seen Figure 3.

Figure 3. The average plasmid loss measured as non-resistant CFU as a percentage counts for the ampicillin resistance of the weak and strong promoter, as well as for chloramphenicol and kanamycin resistance controls. Samples were obtained for spread-plating at 12 hours after incubation start.
From this test, it is evident that antibiotic resistance is reduced for the ampicillin-resistance carrying bacteria, compared to chloramphenicol and kanamycin resistant cultures. This consolidates the hypothesis, that the bacteria transformed with the weak and strong promoter controlling RFP expression lose their plasmids throughout growth. However, it was further observed, that the plasmid loss in bacteria carrying the strong constitutive promoter was markedly higher than for bacteria carrying the weak constitutive promoter. This could be due to the fact that RFP in high concentrations is toxic for the cells, resulting in an increased pressure to lose the plasmid.
When using the promoter to express a gene, the observed plasmid loss should be taken into account, and compensated for, e.g. by continuous supply of antibiotic or substitution of the ampicillin resistance cassette.
|
East Chapel Hill 2018 : Measuring strength of J23102 in novel chassis |
|
[http://2019.igem.org/Team:Tacoma_RAINmakers Tacoma RAINmakers 2019] |
Tacoma RAINmakers 2019: Relative Strength of Anderson Promoters
We collected characterization data on the relative RFP expression rates of Anderson Promoters BBa_J23100, BBa_J23101, BBa_J23102, BBa_J23105, BBa_J23106, and BBa_J23112 in the RFP expression plasmid BBa_J61002 (samples provided in the 2019 iGEM Distribution Kit). 
|
|
•••••
University of Texas at Austin iGEM 2019 |
Characterization of the Anderson series through burden monitoring by the University of Texas at Austin's 2019 iGEM teamDescriptionOur team transformed the Anderson series of RFP reporters (J23101, J23113, J23104, J23107, J23117 in pSBC13 backbone) into our constitutive GFP burden monitor E. coli strain and measured the relative burden of each part. They contained a constitutive GFP sequence in the genome which serves as a way to measure the constructs’ ribosome allocation. Our results show a certain reduction ingrowth rate for each part as a result of ribosome misallocation away from the genome and towards the plasmid containing the construct. The promoter strengths associated with each RFP reporter construct shows that parts with stronger promoters express less GFP and have a reduced growth rate when compared to the constructs containing weaker promoters. The promoters associated with these RFP reporters were used to create a series of BFP reporters, also transformed into our GFP burden monitor strain, and create the regression on the figure below. For more information on characterization of these parts through burden monitoring and evolutionary stability experiments, visit our team’s wiki page: [1] |