Difference between revisions of "Part:BBa K2656102"
Carolinarp (Talk | contribs) |
Carolinarp (Talk | contribs) |
||
| Line 14: | Line 14: | ||
</ul> | </ul> | ||
</html> | </html> | ||
| + | |||
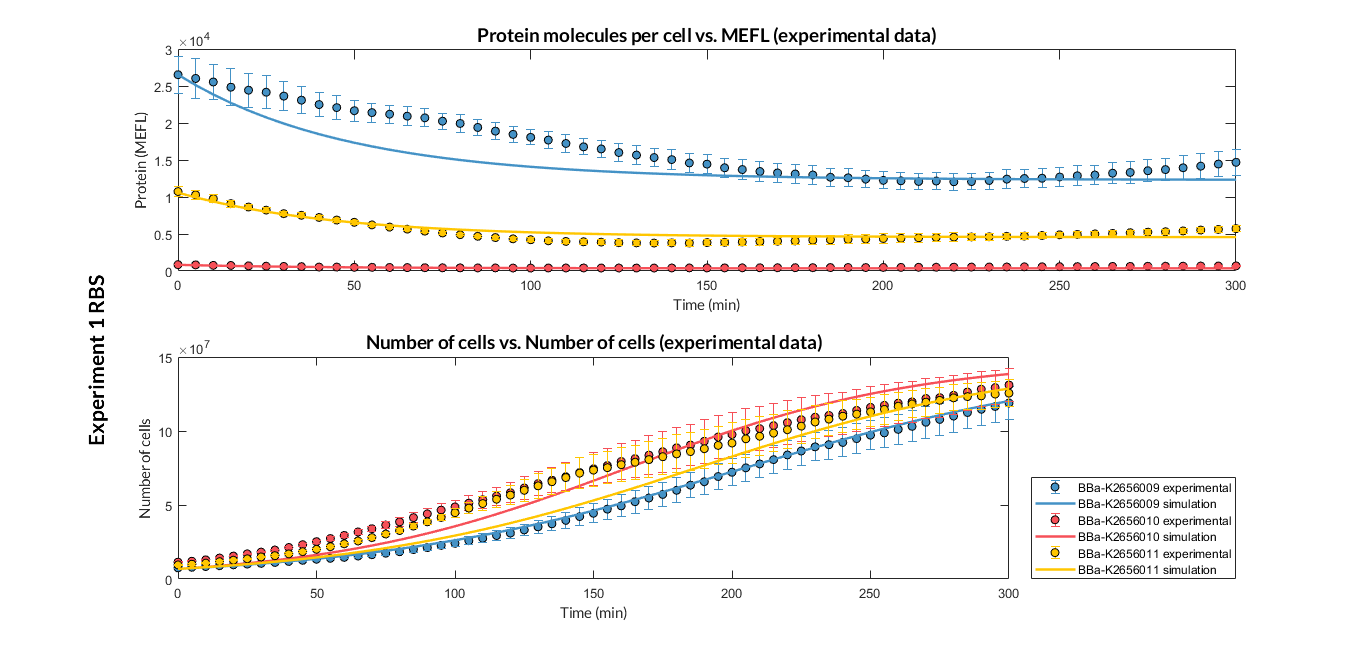
| + | By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model] and the results of the next graphs have been obtained after an optimization and decision-making process, in which the optimal parameters have been selected and the constituent model for each objective has been simulated and compared with the experimental data. | ||
| + | |||
| + | [[File:T--Valencia_UPV--optimization_exp1_RBS_graphUPV2018.png|900px|thumb|none|alt=RBS experiment 1.|Figure 1. RBS expression experiment with K2656009, K26560010 and K2656011 RBS basic parts]] | ||
| + | |||
| + | |||
| + | {|class='wikitable' | ||
| + | |colspan=4|Table 1. Optimized parameters for the BBa_K2656010 RBS. | ||
| + | |- | ||
| + | |'''Parameter''' | ||
| + | |'''Value''' | ||
| + | |- | ||
| + | |Translation rate p | ||
| + | |p = 0.01 min-1 | ||
| + | |- | ||
| + | |Dilution rate μ | ||
| + | | μ = 0.01641 min-1 | ||
| + | | PoI degradation rate | ||
| + | | RBS Experiment 1 dp = 0.0058 min-1 | ||
| + | |} | ||
Revision as of 21:49, 14 October 2018
sfGFP transcriptional unit 3
Constitutive expressed transcriptional unit assembled with a one-pot [http://2018.igem.org/Team:Valencia_UPV/Design Level 1] Golden Gate reaction using BsaI type IIS endonuclease.
This transcriptional unit is composed of the following standardized parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]:
- BBa_K2656004: the J23106 constitutive promoter in its Golden Braid compatible version
- BBa_K2656010: the B0032 weak ribosome biding site in its Golden Braid compatible version
- BBa_K2656013: the BBa_I746916 sfGFP sequence in its Golden Braid standardized version
- Terminator BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model] and the results of the next graphs have been obtained after an optimization and decision-making process, in which the optimal parameters have been selected and the constituent model for each objective has been simulated and compared with the experimental data.
| Table 1. Optimized parameters for the BBa_K2656010 RBS. | |||
| Parameter | Value | ||
| Translation rate p | p = 0.01 min-1 | ||
| Dilution rate μ | μ = 0.01641 min-1 | PoI degradation rate | RBS Experiment 1 dp = 0.0058 min-1 |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 11
Illegal NheI site found at 34 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 76