Difference between revisions of "Part:BBa E1010"
Herlohuang (Talk | contribs) |
Herlohuang (Talk | contribs) |
||
| Line 73: | Line 73: | ||
</table> | </table> | ||
| + | ===Contribution3=== | ||
| + | Part name:<partinfo>BBa_K2382014</partinfo> | ||
| + | <br> | ||
| + | Group: iGEM17_CSMU_NCHU_Taiwan 2017 | ||
| + | <br> | ||
| + | Author: TING-YU LIN | ||
| + | <br> | ||
| + | Summary: This part encodes a RFP that has the same amino acids as BBa_E1010. The barcode of BBa_E1010 is removed, so this part does not contain stop codon. Therefore, future iGEM teams could fuse other protein at the C terminal of this RFP. For more, we use the IDT Codon Optimization Tool(http://sg.idtdna.com/CodonOpt) to optimize the sequence and make it more suit in E.coli K12 | ||
| + | <br> | ||
| + | Documentation: | ||
| + | ===<span class='h3bb'>Sequence and Features</span>=== | ||
| + | <partinfo>BBa_K2382014 SequenceAndFeatures</partinfo> | ||
===Contribution2=== | ===Contribution2=== | ||
| + | Part name:<partinfo>BBa_K2382013</partinfo> | ||
| + | <br> | ||
Group: iGEM17_CSMU_NCHU_Taiwan 2017 | Group: iGEM17_CSMU_NCHU_Taiwan 2017 | ||
<br> | <br> | ||
Author: SHAO-CHI LO | Author: SHAO-CHI LO | ||
<br> | <br> | ||
| − | Summary: We add a His Tag at the end. Therefore, a fusion protein with this part may have red color and the ability to be purified easily. | + | Summary: We add a His Tag at the end. Therefore, a fusion protein with this part may have red color and the ability to be purified easily. |
<br> | <br> | ||
Documentation: | Documentation: | ||
===<span class='h3bb'>Sequence and Features</span>=== | ===<span class='h3bb'>Sequence and Features</span>=== | ||
<partinfo>BBa_K2382013 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2382013 SequenceAndFeatures</partinfo> | ||
| + | |||
Revision as of 06:17, 30 October 2017
**highly** engineered mutant of red fluorescent protein from Discosoma striata (coral)
monomeric RFP: Red Fluorescent Protein. Excitation peak: 584 nm Emission peak: 607 nm
Usage and Biology
Robert E. Campbell started with Discosoma RFP (DsRed) and evolved a faster folding, monomeric variant. See paper listed in source. Codon optimized for expression in bacteria (?? DE)
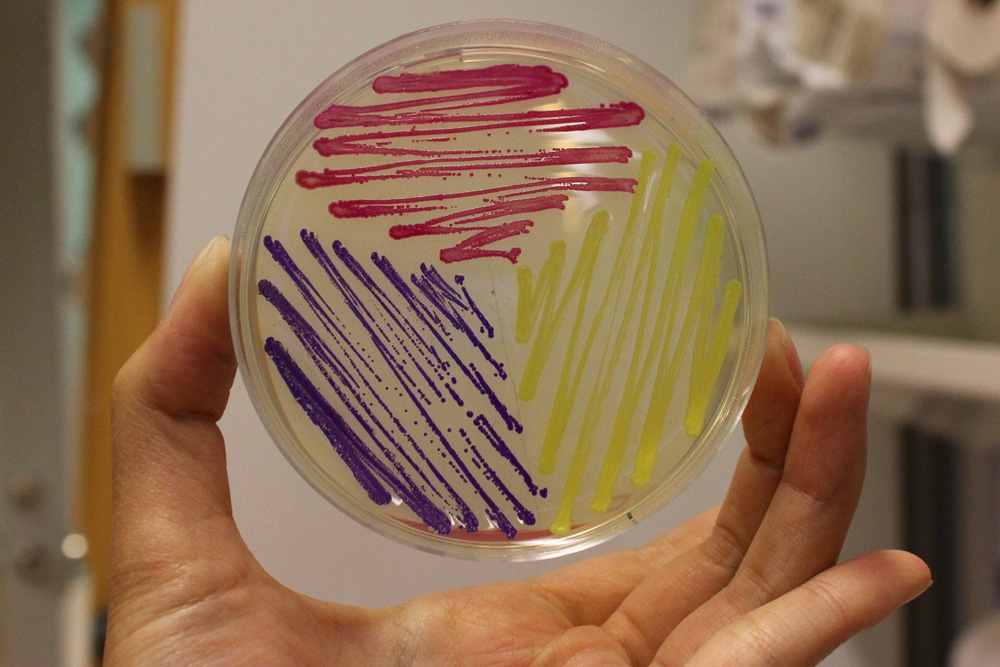
iGEM11_Uppsala-Sweden: Expression of chromoproteins. The images above show E coli constitutively expressing amilCP BBa_K592009 (blue), amilGFP BBa_K592010 (yellow) and RFP BBa_E1010 (red).
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989004".
Contribution
Group: Hong Kong-CUHK iGEM 2017
Author: Yuet Ching Lin
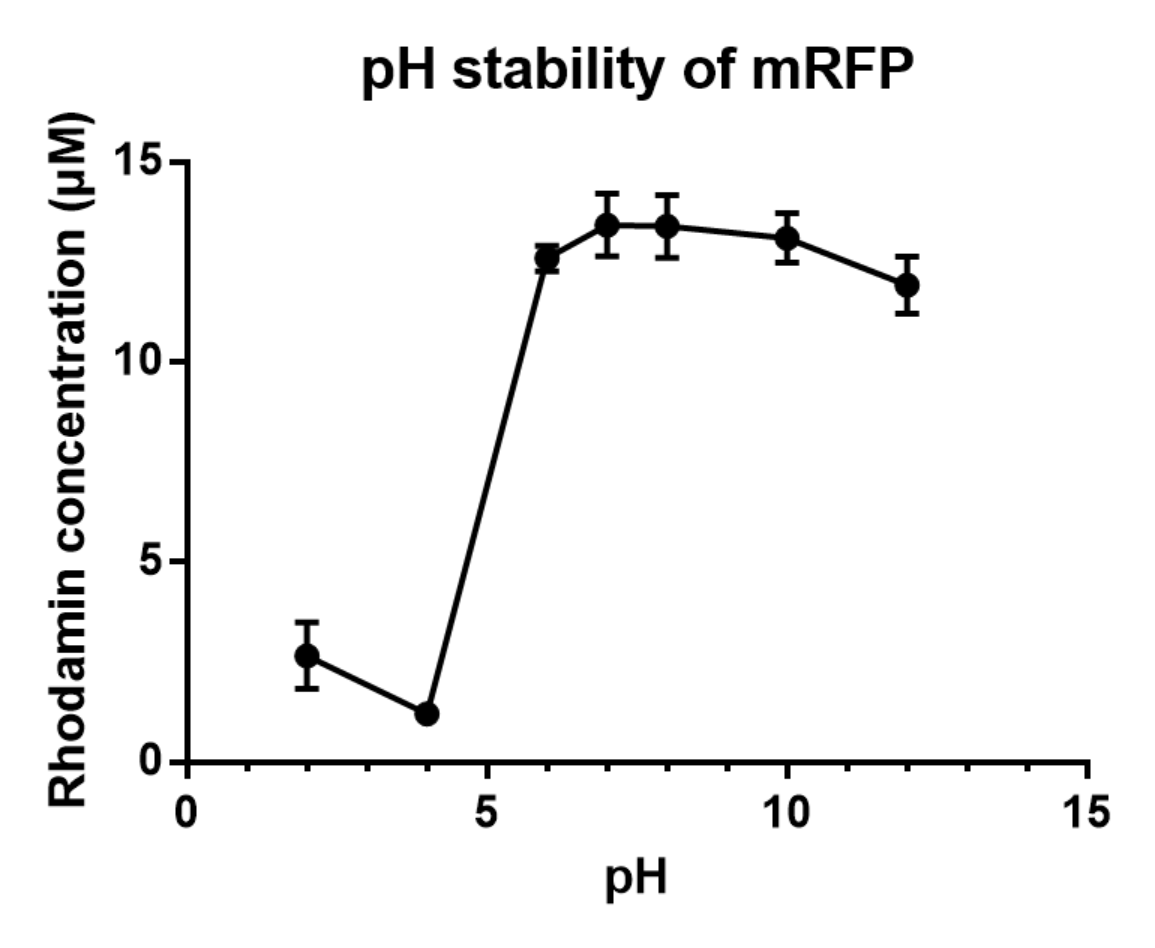
Summary: We measured the fluorescent signal of mRFP in buffers with different pH.
Documentation:
Charaterization of mRFP pH stabillity:
We grew C41 bacteria with parts BBa_J61002 in 2XYT for 24 hours. After purifying the mRFP by Ion Exchange Chromatography and Hydrophobic Interaction Chromatography, we measured the fluoresece (ex ,em ) of purified mRFP, which is diluted to 10µg/100µl (total 200µl) in triplicates, into different buffers (ranges from pH2 to pH12; Volume of mRFP:buffer = 1:1.8). To facilitate reproducibility of the experiment, we correlated the relative fluorescent intensity to an absolute fluorophore concentration by referring it to a standard curve of the fluorophores(Rhodamine) using the interlab study protocol. The result shows that the stability drops dramatically in pH condition below 6 and relatively stable in pH 6-10.
| Measurement Type | Fluorescence |
| Microplate name | COSTAR 96 |
| Scan mode | orbital averaging |
| Scan diameter [nm] | 3 |
| Excitation | 550-20 |
| Emission | 605-40 |
| Dichronic filter | auto 572.5 |
| Gain | 500 |
| Focal height [nm] | 9 |
Contribution3
Part name:BBa_K2382014
Group: iGEM17_CSMU_NCHU_Taiwan 2017
Author: TING-YU LIN
Summary: This part encodes a RFP that has the same amino acids as BBa_E1010. The barcode of BBa_E1010 is removed, so this part does not contain stop codon. Therefore, future iGEM teams could fuse other protein at the C terminal of this RFP. For more, we use the IDT Codon Optimization Tool(http://sg.idtdna.com/CodonOpt) to optimize the sequence and make it more suit in E.coli K12
Documentation:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 555
- 1000COMPATIBLE WITH RFC[1000]
Contribution2
Part name:BBa_K2382013
Group: iGEM17_CSMU_NCHU_Taiwan 2017
Author: SHAO-CHI LO
Summary: We add a His Tag at the end. Therefore, a fusion protein with this part may have red color and the ability to be purified easily.
Documentation:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 606
- 1000COMPATIBLE WITH RFC[1000]
|};
Parts table
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||