Difference between revisions of "Part:BBa K1041000"
(→Characterisation) |
(→Characterisation) |
||
| Line 25: | Line 25: | ||
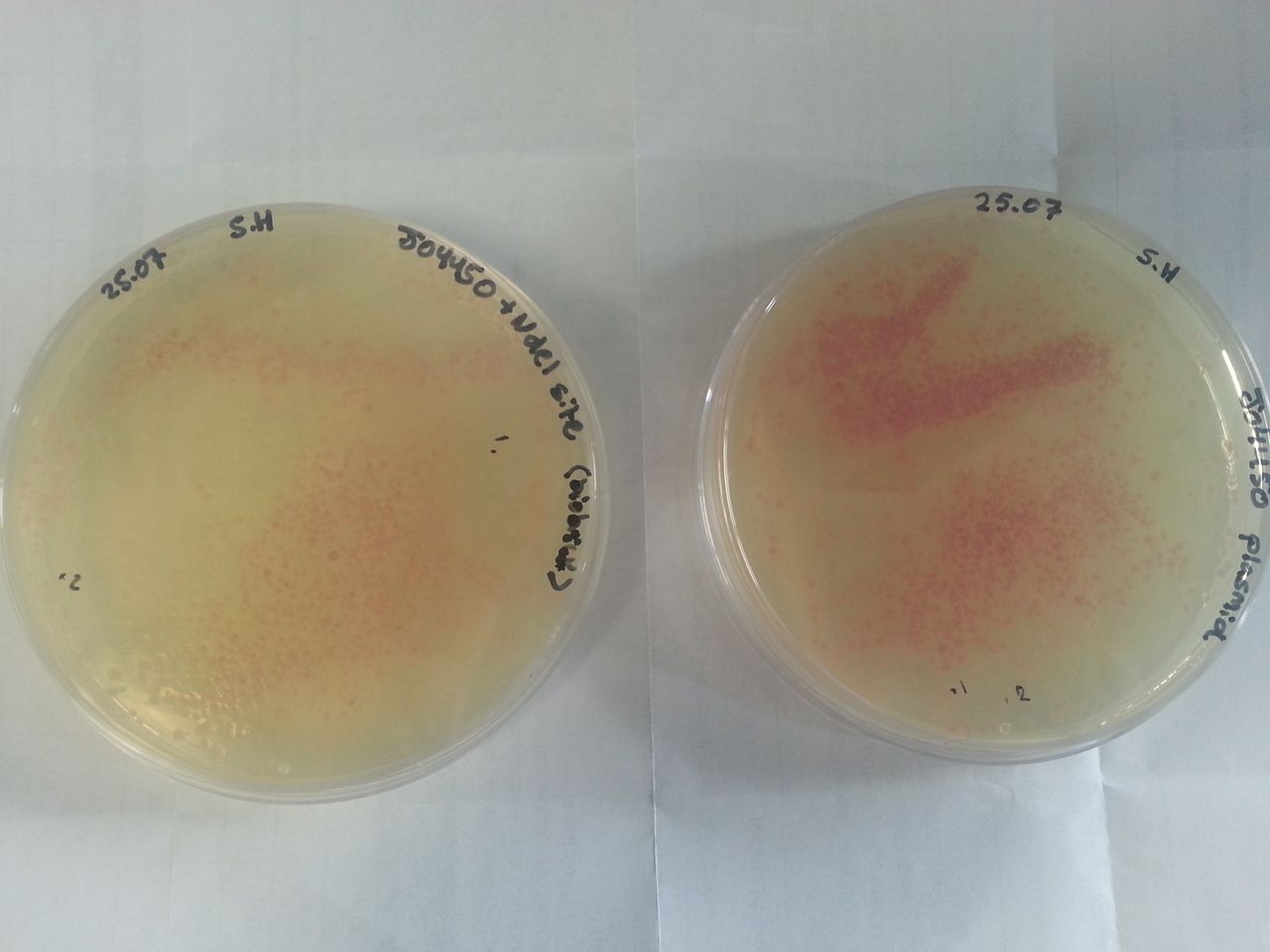
[[Image:WP 000051.jpg|thumb|Fig 1: Plates containing (left) BBa_K1041000 and (right) BBa_J04450 visualized under non-UV lightbox]] | [[Image:WP 000051.jpg|thumb|Fig 1: Plates containing (left) BBa_K1041000 and (right) BBa_J04450 visualized under non-UV lightbox]] | ||
| − | + | <br> | |
===Sequencing=== | ===Sequencing=== | ||
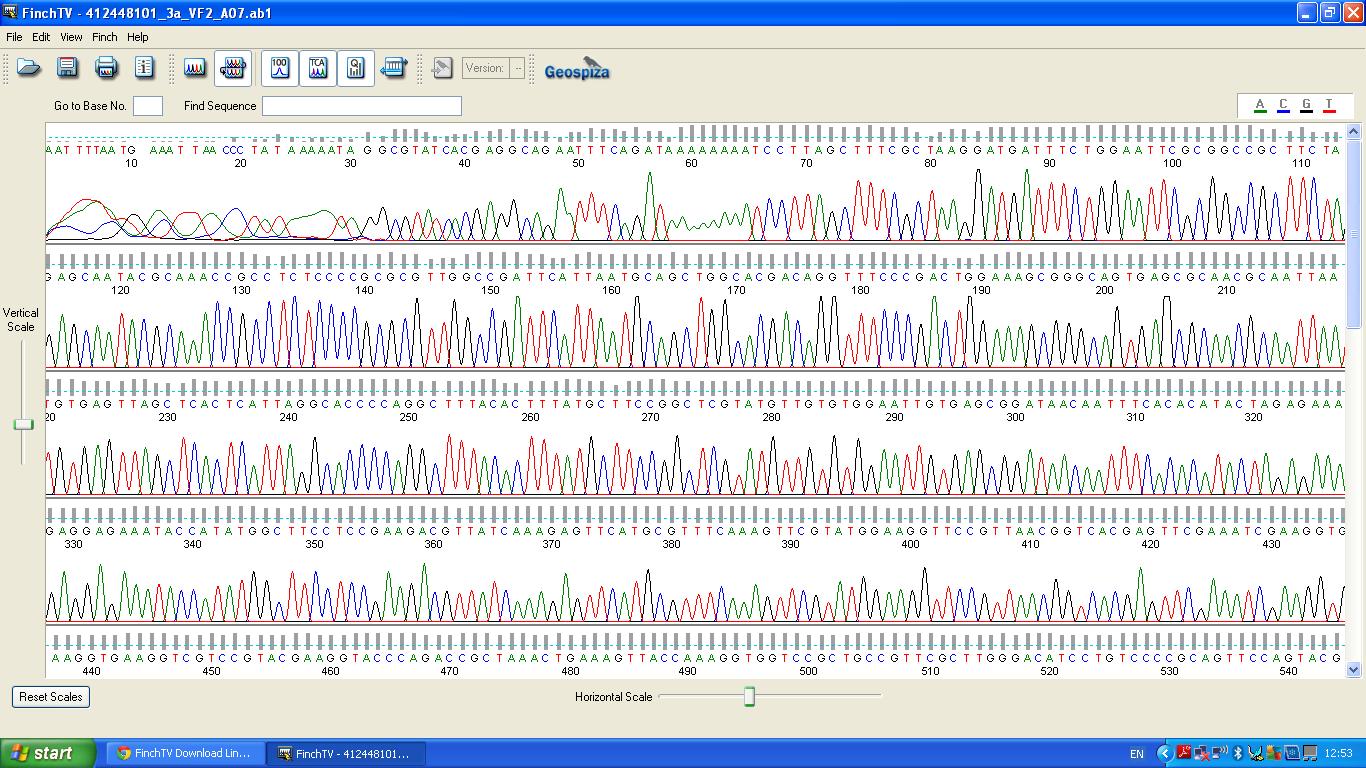
The biobrick was sent off to a company for sequencing. | The biobrick was sent off to a company for sequencing. | ||
| Line 32: | Line 32: | ||
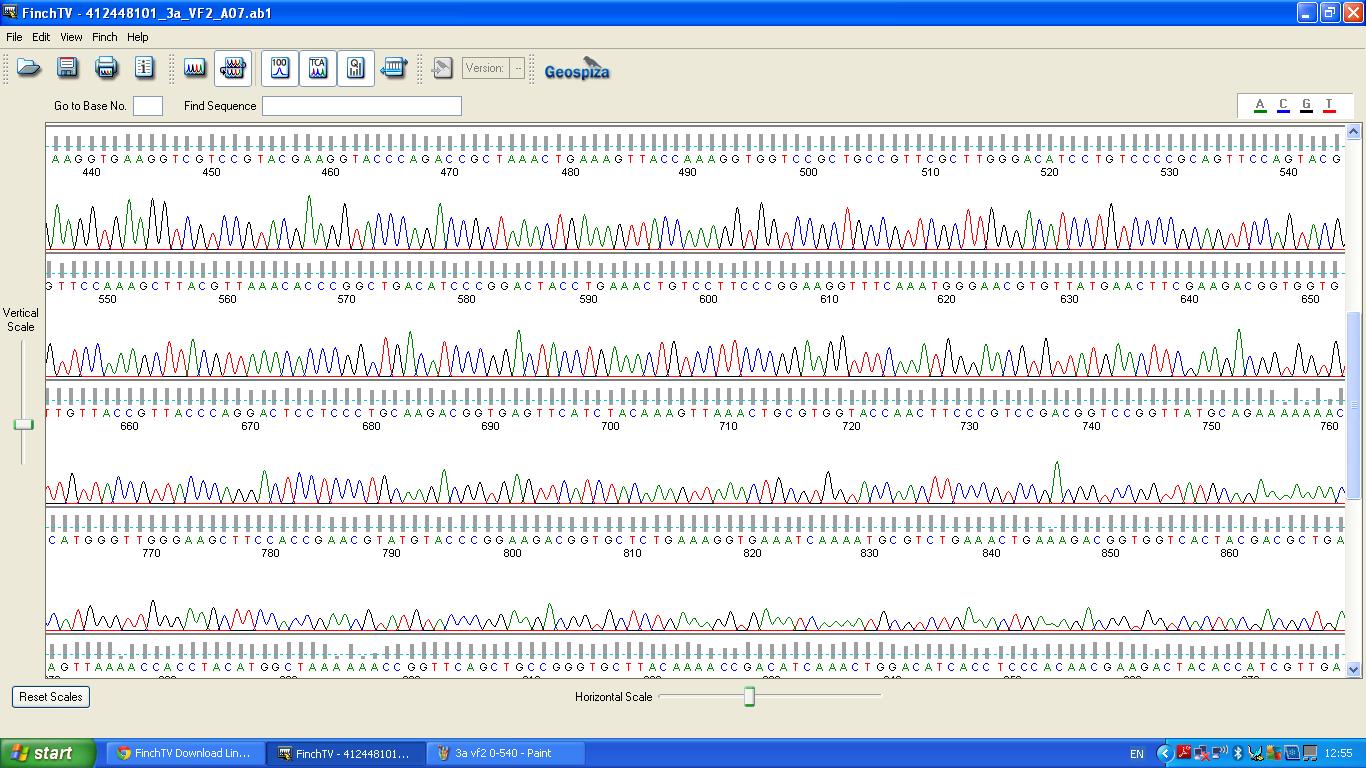
[[image:3a vf2 440-860.JPG|thumb|left|K1041000 sequencing data part 2 ]] | [[image:3a vf2 440-860.JPG|thumb|left|K1041000 sequencing data part 2 ]] | ||
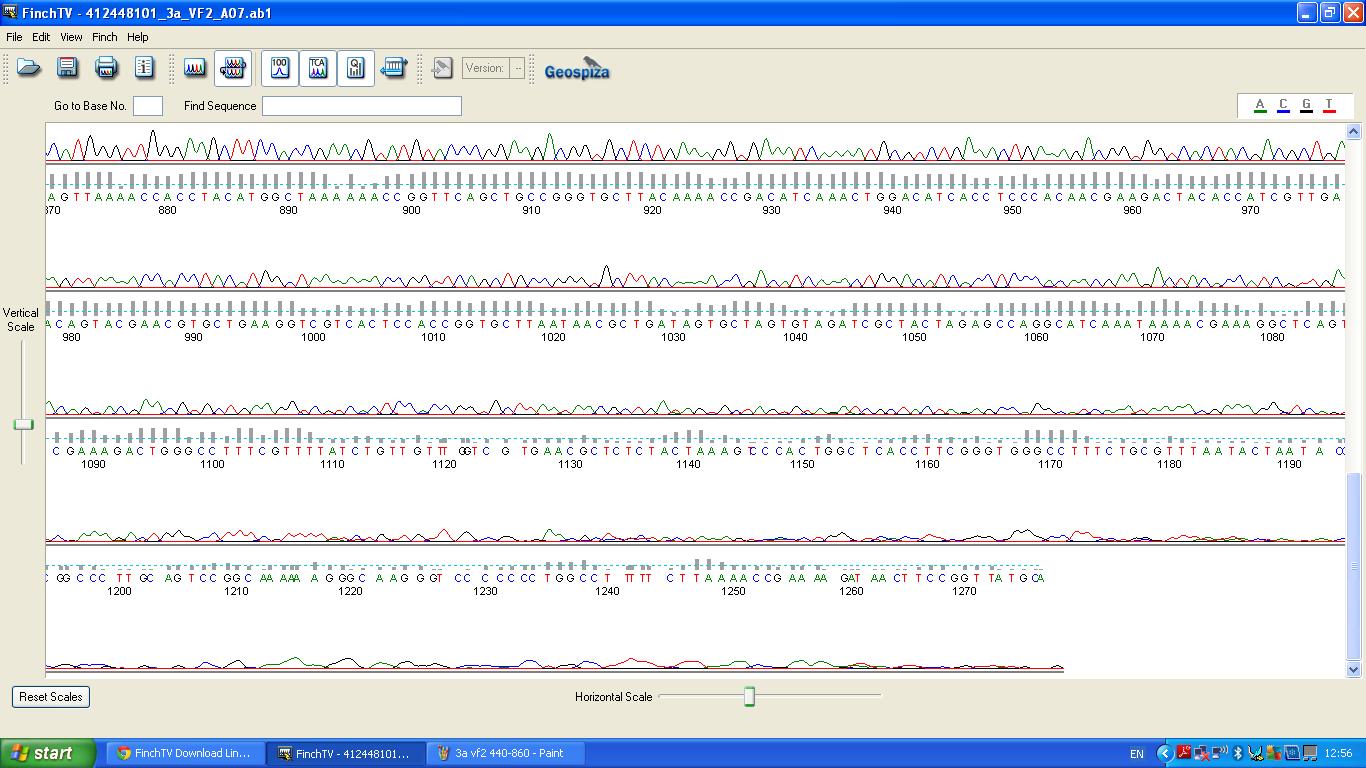
[[image:3a vf2 860-.JPG|thumb|left|K1041000 sequencing data part 3]] | [[image:3a vf2 860-.JPG|thumb|left|K1041000 sequencing data part 3]] | ||
| − | + | <br> | |
===BLAST Analysis=== | ===BLAST Analysis=== | ||
The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence ''fig.2'' | The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence ''fig.2'' | ||
Revision as of 10:56, 17 September 2013
RFP Coding Device
Team NRP-UEA (2013) used mutagenesis to add a Nde1 site at the start of the RFP coding region of the biobrick BBa_J04450. This enables a restriction digest with Nde1 to be performed allowing the RFP coding region to be excised from the plasmid.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 781
Illegal AgeI site found at 893 - 1000COMPATIBLE WITH RFC[1000]
Composite part of the following BioBricks:
- BBa_R0010: Promoter (lacI regulated)
- BBa_B0034: RBS (Elowitz 1999)
- BBa_E1010: Red Fluorescent Protein from Discosoma striata
- BBa_B0015: Double Terminator
Characterisation
Characterisation of this biobrick involved comparisons with the original Bba_J04450 biobrick by performing transformations, restriction digests and BLAST analysis. We also had our biobrick sequenced.
Transformation
Parts BBa_K1041000 and BBa_J04450 were transformed into Alpha-Select competent E.Coli cells and plated onto LB Agar plates which contained the antibiotic ampicillin. Both plates have colonies that appear red under natural light fig.1. Therefore the addition of a NdeI site has not effected the ability for the RFP gene to be transcribed.
Sequencing
The biobrick was sent off to a company for sequencing.
BLAST Analysis
The data we recieved back from the sequencing company was aligned using BLAST with the expected DNA sequence fig.2