Difference between revisions of "Protein coding sequences/Biosynthesis/Bisphenol A"
(New page: < Back to Biosynthesis Protein coding sequences <html> <style> #assembly_plasmid_table {margin-left:auto;margin-right:auto; border: 3px solid #44...) |
|||
| Line 9: | Line 9: | ||
</html> | </html> | ||
| − | Bisphenol A is a toxin that has been shown to leech from certain types of plastic. Studies have shown this chemical to have detrimental effects in animal studies and is very likely to be harmful to humans as well. The following bisphenol A degradation protein coding sequences are from ''Sphingomonas bisphenolicum'' and may aid in the remediation of bisphenol A contamination. | + | Bisphenol A is a toxin that has been shown to leech from certain types of plastic. Studies have shown this chemical to have detrimental effects in animal studies and is very likely to be harmful to humans as well. The following bisphenol A degradation protein coding sequences are from ''Sphingomonas bisphenolicum'' AO1 and may aid in the remediation of bisphenol A contamination. |
<parttable>bisphenol_degradation_protein_coding_sequence</parttable> | <parttable>bisphenol_degradation_protein_coding_sequence</parttable> | ||
| Line 19: | Line 19: | ||
|Jason Gardiner and the [http://2008.igem.org/Team:The_University_of_Alberta 2008 University of Alberta iGEM team] designed a set of parts for degradation of bisphenol A. | |Jason Gardiner and the [http://2008.igem.org/Team:The_University_of_Alberta 2008 University of Alberta iGEM team] designed a set of parts for degradation of bisphenol A. | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | {| | ||
| + | |[[Image:Bielefeld-Germany2011-Logo auf weiss.png|50px]] | ||
| + | |In 2011, the iGEM team from [http://2011.igem.org/Team:Bielefeld-Germany Bielefeld University] could show that fusing BisdA and BisdB together to one protein leeds to an improvement of the bisphenol A degradation. | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ===Background of bisphenol A degradation=== | ||
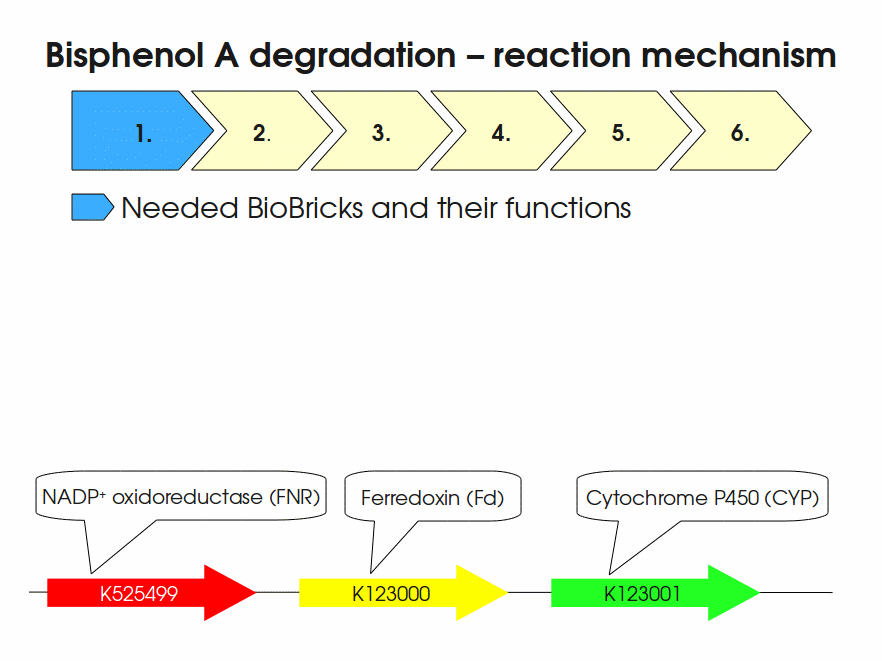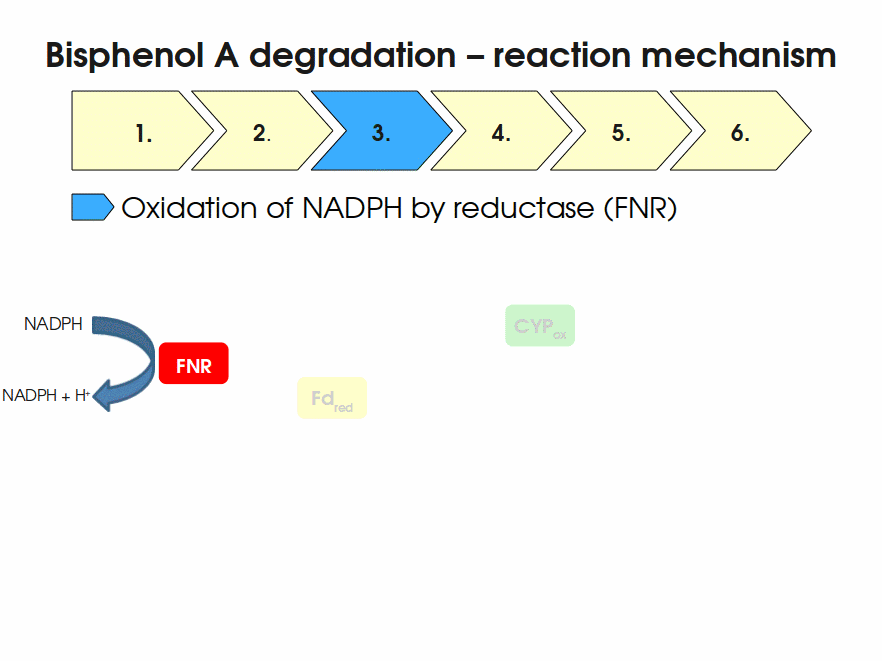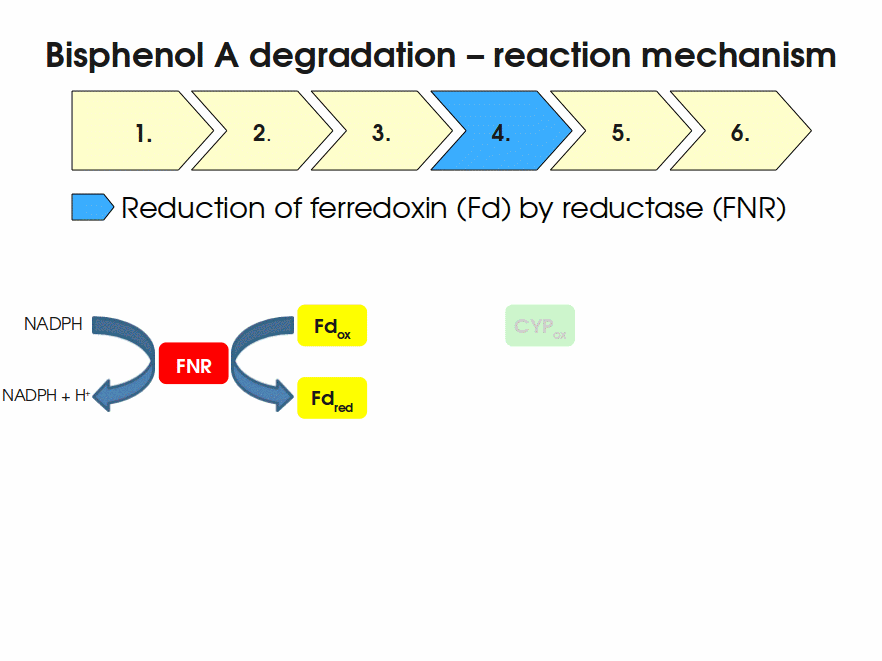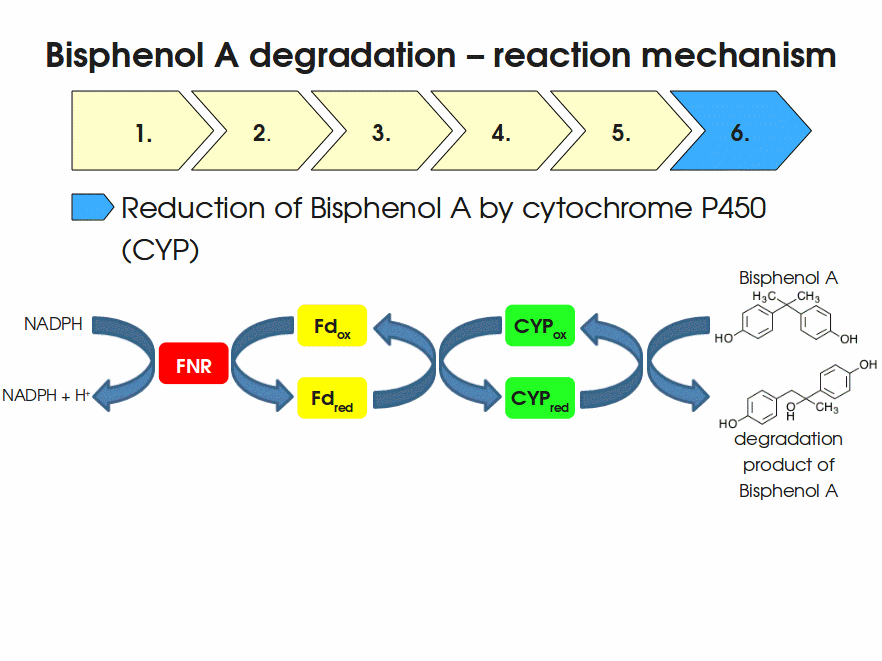
| + | Bisphenol A is mainly hydroxylated into the products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol. In ''S. bisphenolicum'' AO1, a total of three genes are responsible for this BPA hydroxylation: a cytochrome P450 (CYP, ''bisdB''), a ferredoxin (Fd, ''bisdA'') and a ferredoxin-NAD<sup>+</sup> oxidoreductase (FNR) <cite>Sasaki05a</cite>. The three gene products act together to reduce BPA while oxidizing NADH + H<sup>+</sup>. The cytochrome P450 (BisdB) reduces the BPA and is oxidized during this reaction. BisdB in its oxidized status is reduced by the ferredoxin (BisdA) so it can reduce BPA again. The oxidized BisdA is reduced by a ferredoxin-NAD<sup>+</sup> oxidoreductase consuming NADH + H<sup>+</sup> so the BPA degradation can continue <cite>Sasaki05a</cite>. This electron transport chain between the three enzymes involved in BPA degradation and the BioBricks needed to enable this reaction ''in vivo'' and ''in vitro'' are shown in the following figure (please have some patience, it's an animated .gif file): | ||
| + | |||
| + | [[Image:Bielefeld-Germany2011-BPAdegrad2.gif|center|700px|thumb|'''Fig. 1: Animation of proposed reaction mechanism of bisphenol A hydroxylation by the involved enzymes FNR (<partinfo>K525499</partinfo>, Fd (BisdA, <partinfo>K123000</partinfo>) and CYP (BisdB, <partinfo>K123001</partinfo>)''']] | ||
| + | |||
| + | |||
===References=== | ===References=== | ||
Revision as of 12:55, 17 September 2011
< Back to Biosynthesis Protein coding sequences
Bisphenol A is a toxin that has been shown to leech from certain types of plastic. Studies have shown this chemical to have detrimental effects in animal studies and is very likely to be harmful to humans as well. The following bisphenol A degradation protein coding sequences are from Sphingomonas bisphenolicum AO1 and may aid in the remediation of bisphenol A contamination.
| Name | Protein | Description | Direction | Uniprot | KEGG | E.C. | Substrate | Product | Length | Status |
|---|---|---|---|---|---|---|---|---|---|---|
| BBa_K525515 | Fusion protein of BisdA and BisdB | 1602 | In stock | |||||||
| BBa_K525517 | Fusion protein of BisdA and BisdB behind constitutive promoter | 1663 | In stock | |||||||
| BBa_K123000 | BisdA | 330 | It's complicated | |||||||
| BBa_K123001 | BisdB | 1284 | It's complicated | |||||||
| BBa_K525007 | Fusion protein of S-Layer SgsE with T7/lac and BisdA and BisdB | 3999 | Not in stock | |||||||
| BBa_K525008 | Fusion protein of S-Layer SbpA with T7/lac and BisdA and BisdB | 4791 | Not in stock | |||||||
| BBa_K525582 | Fusion protein of NADP+ Oxidoreductase and BisdA and BisdB, polycistronic | 2448 | Not in stock | |||||||
| BBa_K525562 | Fusion protein of NADP+ Oxidoreductase and BisdA and BisdB with middle strong promoter,RBS | 2410 | Not in stock |
| Jason Gardiner and the [http://2008.igem.org/Team:The_University_of_Alberta 2008 University of Alberta iGEM team] designed a set of parts for degradation of bisphenol A. |
Background of bisphenol A degradation
Bisphenol A is mainly hydroxylated into the products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol. In S. bisphenolicum AO1, a total of three genes are responsible for this BPA hydroxylation: a cytochrome P450 (CYP, bisdB), a ferredoxin (Fd, bisdA) and a ferredoxin-NAD+ oxidoreductase (FNR) Sasaki05a. The three gene products act together to reduce BPA while oxidizing NADH + H+. The cytochrome P450 (BisdB) reduces the BPA and is oxidized during this reaction. BisdB in its oxidized status is reduced by the ferredoxin (BisdA) so it can reduce BPA again. The oxidized BisdA is reduced by a ferredoxin-NAD+ oxidoreductase consuming NADH + H+ so the BPA degradation can continue Sasaki05a. This electron transport chain between the three enzymes involved in BPA degradation and the BioBricks needed to enable this reaction in vivo and in vitro are shown in the following figure (please have some patience, it's an animated .gif file):
References
<biblio>
- Sasaki pmid=18492046
- Zhang pmid=17291567
- Nakajima pmid=12354921
- Sasaki05a pmid=16332782
- Sasaki05b pmid=15865158
- Kang02 pmid=12202920
- Kang04 pmid=15242458
</biblio>
