Difference between revisions of "Part:BBa K4162117"
m |
m |
||
| Line 22: | Line 22: | ||
====Agarose gel electrophoresis==== | ====Agarose gel electrophoresis==== | ||
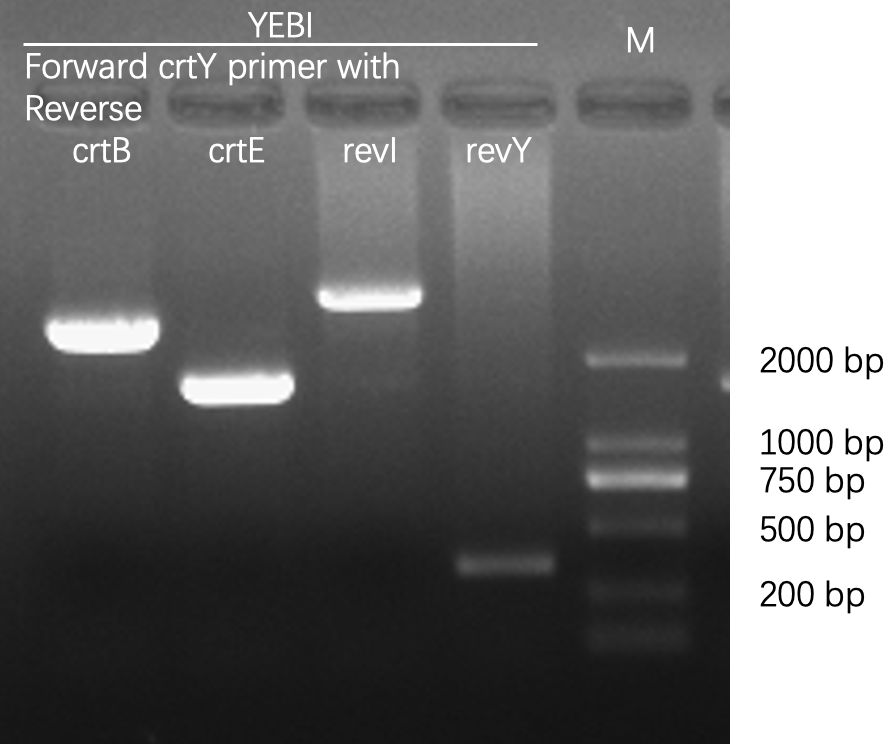
| − | [[File:T--Fudan--Agarose gel electrophoresis--crtYEBI.png|400px|thumb|none|'''Figure 1. Agarose gel electrophoresis of PCR products, amplified from bacterial colonies/cultures.''' The first lane was loaded with D2000 DNA ladder whose sizes were marked on the image. We chose Taq DNA polymerase for its low cost and high reliability, and we designed forward and reverse primers for each carotene synthesis enzyme (crt for short). The PCR reaction was composed of 2 μL 10x Taq polymerase buffer, 16 μL | + | [[File:T--Fudan--Agarose gel electrophoresis--crtYEBI.png|400px|thumb|none|'''Figure 1. Agarose gel electrophoresis of PCR products, amplified from bacterial colonies/cultures.''' The first lane was loaded with D2000 DNA ladder whose sizes were marked on the image. We chose Taq DNA polymerase for its low cost and high reliability, and we designed forward and reverse primers for each carotene synthesis enzyme (crt for short). The PCR reaction was composed of 2 μL 10x Taq polymerase buffer, 16 μL H<sub>2</sub>O, 0.5 μL Taq polymerase, 0.5 μL dNTP (10 mM each), 0.5 μL forward primer (10 mM), 0.5 μL reverse primer (10 mM), and 1 μL bacterial culture or 1μL colony. Using the same forward primer, and different reverse primers, we were able to detect the composition of various crt genes. After PCR, the correct bacterial clones were sent for Sanger sequencing. Once verified, these clones would be used for further experiments. The sequences of primers are: > 5-crtY 5-ATGCAACCGCATTATGATCTGATTC-3; > rev320crtB 5-CCTTCCAGATGATCAAACGCGTAAG-3; > rev320crtE 5-ATGAGAATGAATGGTAGGGCGTC-3; > rev320crtI 5-GGATTAAACTGCTGAATCTGCGCTTC-3; > rev320crtY 5-CCGCGGTATCCATCCACAAG-3.]] |
====Successful protein expression==== | ====Successful protein expression==== | ||
Revision as of 09:30, 12 October 2022
ribozyme+RBS+CDS module: crtYEBI
Introduction
This biobrick was created through overlapping PCR of BBa_K4162020(ribozyme+J6_RBS+crtY), BBa_K4162010(ribozyme+T7_RBS+crtE), BBa_K4162013(ribozyme+T7_RBS+crtB) and BBa_K4162016(ribozyme+T7_RBS+crtI). These genes are a part of the carotenoid biosynthesis pathway and together, this biobrick converts farnesyl pyrophosphate to β-carotene. In this part, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Thus, self-interaction of the polycistron can be avoid, and each individual cistron containing RBS+CDS can initiate translation with comparable efficiency.
Comparing to BBa_K4162021, we use J6_RBS rather T7_RBS to drive the translation crtY. J6_RBS is weaker than T7_RBS, which means less chance for ribosomes to start the translation. Reduced expression of crtY may help the expression of crtE, which is obscure for BBa_K4162021 even under IPTG induction.
Contents
Usage and Biology
We transfected this biobrick into E. coli to build single-cell factory for β-carotene production. Coding sequences of crtYEBI are separated by ribozyme sequences. In this part, the RBS of crtEBI has equal intensity while the RBS of crtY is significantly weaker than the others. Because crtY catalyzes the last step of the carotenoid reaction chain, we guess the concentration of substrate catalyzed by this enzyme is significantly lower than for the first three steps of the reaction. To avoid the problem of flux imbalance in biosynthesis as well as to reduce unnecessary metabolic stress on cells, we intentionally weakened the RBS intensity of crtY.
Our unsuccessful biobrick BBa_K4162021 supports our guess as well.
Characterization
Agarose gel electrophoresis
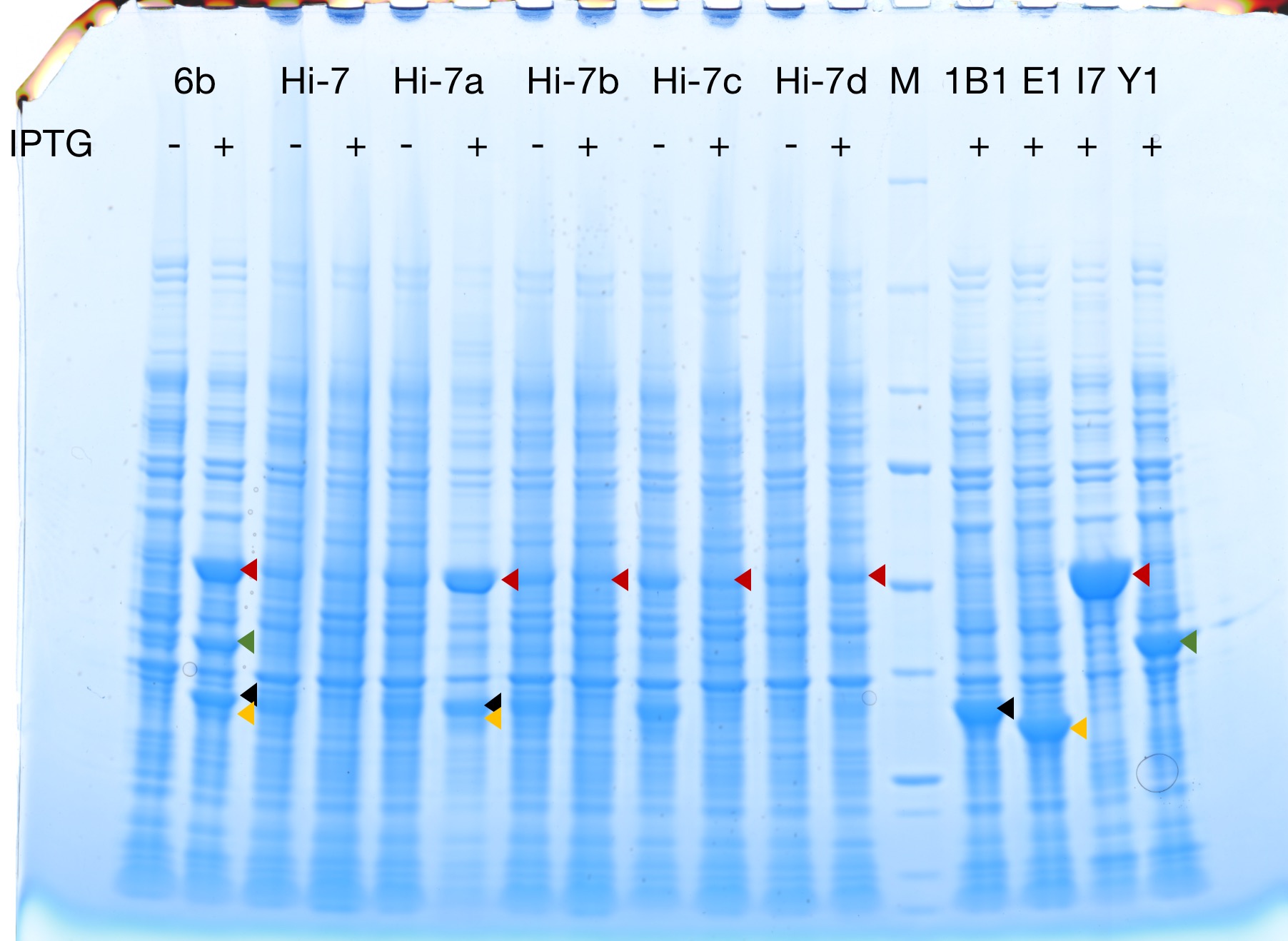
Successful protein expression
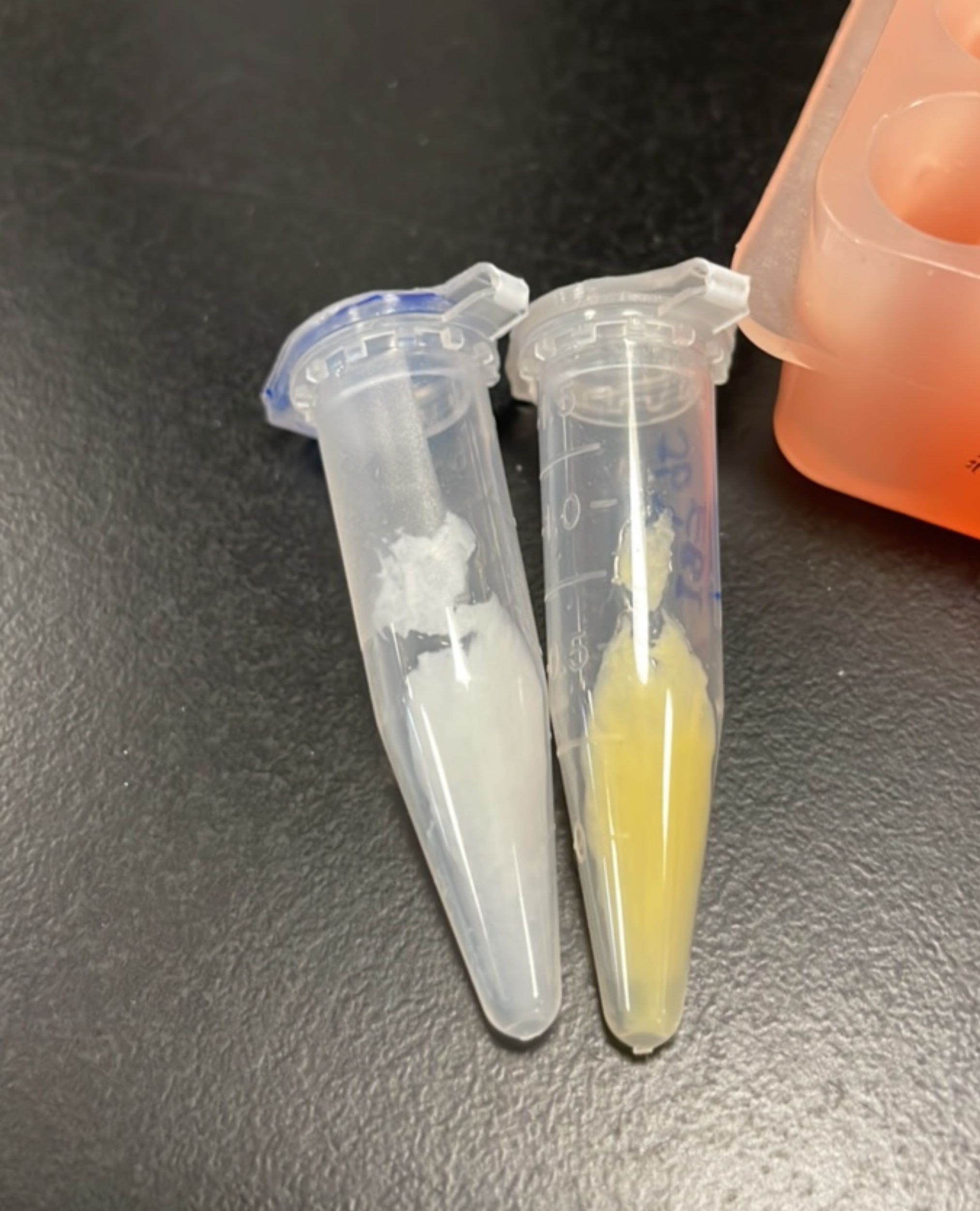
Produce β-carotene
Figures 2 to 4 show that E. coli transfected with this biobrick successfully expressed the target enzyme and yielded β-carotene. In Figure 4, it can be seen that module YEBI corresponds to a darker orange color of the post-centrifugation precipitation compared to module YBEI(BBa_K4162119), characterizing the superior carotenoid yielding ability of module YEBI.

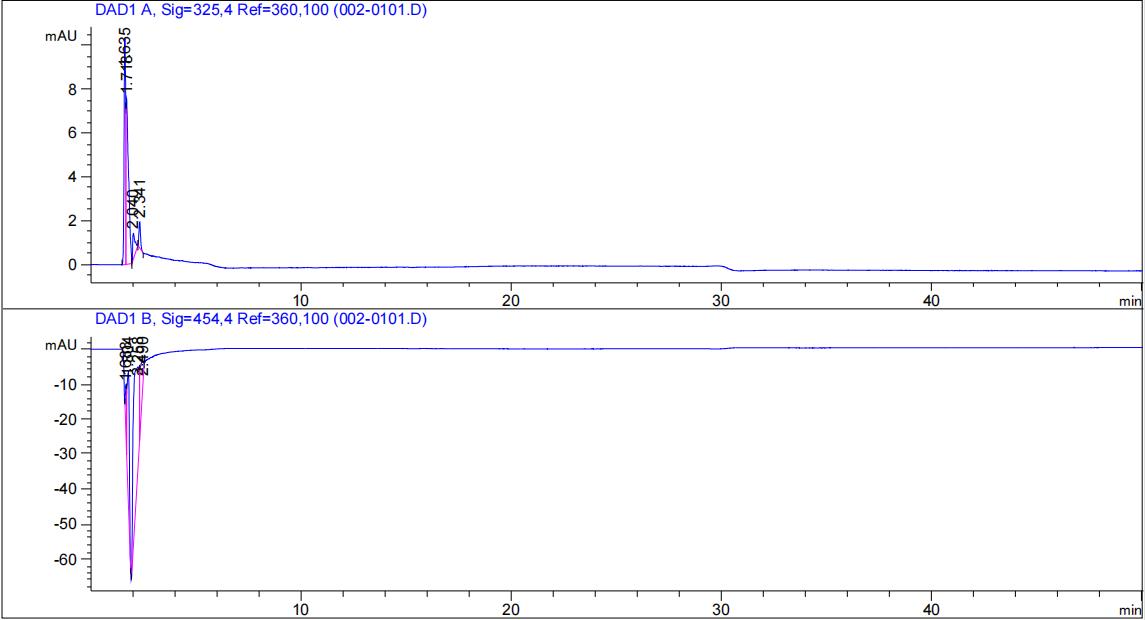
HPLC validation
We used <a href="https://mp.weixin.qq.com/s/81zY_LoMratSEOOy7JUaFw" target="_blank">a previous reported</a> protocol for HPLC: Agilent liquid chromatograph (HPLC-DAD); column C18 (250mm); column temperature 30°C; mobile phase methanol:water = 96:4; flow rate 0.8ml/min; detection wavelength 325 nm and 454 nm.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3425
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2904
Illegal NgoMIV site found at 3034
Illegal AgeI site found at 2062 - 1000COMPATIBLE WITH RFC[1000]