Difference between revisions of "Part:BBa K4150002"
Jacky CHIN (Talk | contribs) |
Jacky CHIN (Talk | contribs) |
||
| Line 21: | Line 21: | ||
<br> | <br> | ||
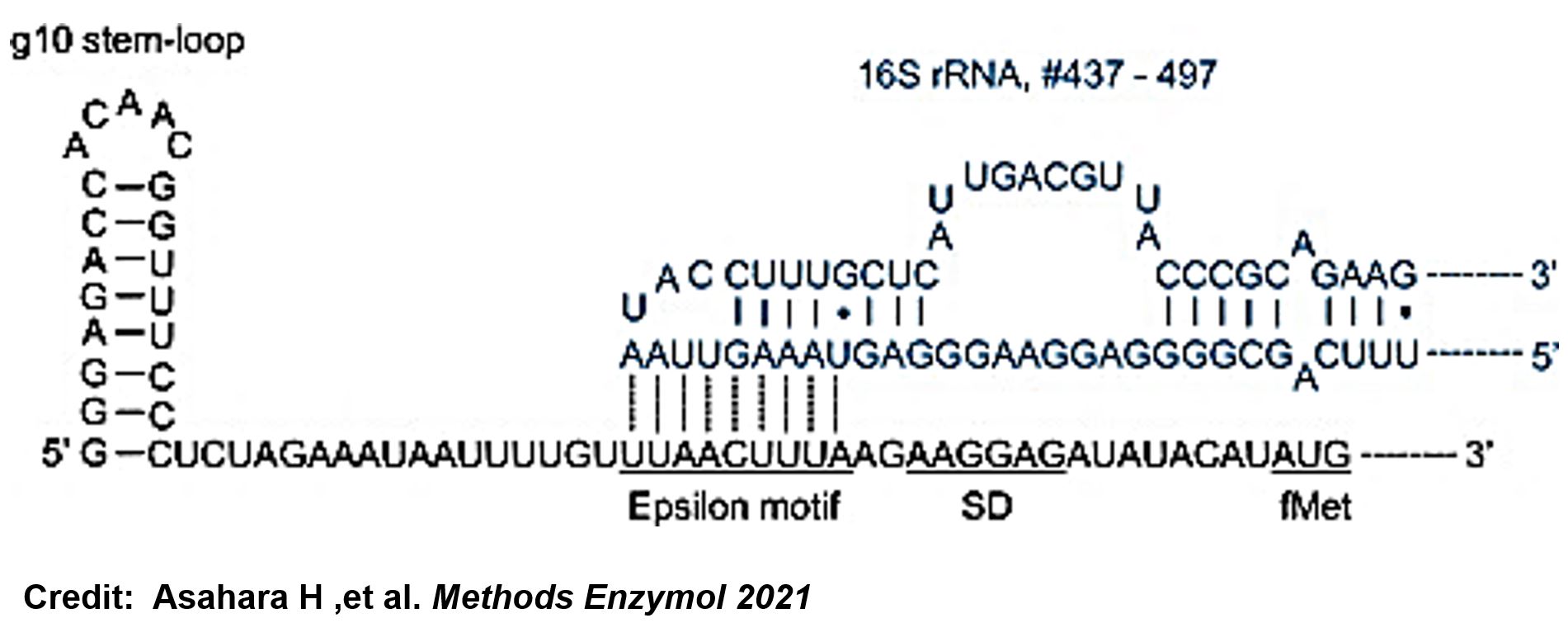
<span style="color:#00000000">This</span>The g10 leader sequence encodes very highly expressed phage proteins, in which 9-base of Epsilon motif exhibits perfect complementary to the 16S RNA of E. coli<ref>Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988 Dec 15;73(1):227-35. doi: 10.1016/0378-1119(88)90329-0. PMID: 3072257.</ref>. This structure connected with the consensus Shine-Dalgarno sequence (SD) can enhance up to 340-fold heterologous gene expression in E. coli<ref>Asahara H, Magnelli P, Shi X, Tuckey C, Zhou Y, Samuelson JC. Guidelines for nucleic acid template design for optimal cell-free protein synthesis using an Escherichia coli reconstituted system or a lysate-based system. Methods Enzymol. 2021;659:351-369. doi: 10.1016/bs.mie.2021.07.005. Epub 2021 Sep 2. PMID: 34752294.</ref>. The g10 leader sequence is also present in several commercially available pET series vectors. | <span style="color:#00000000">This</span>The g10 leader sequence encodes very highly expressed phage proteins, in which 9-base of Epsilon motif exhibits perfect complementary to the 16S RNA of E. coli<ref>Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988 Dec 15;73(1):227-35. doi: 10.1016/0378-1119(88)90329-0. PMID: 3072257.</ref>. This structure connected with the consensus Shine-Dalgarno sequence (SD) can enhance up to 340-fold heterologous gene expression in E. coli<ref>Asahara H, Magnelli P, Shi X, Tuckey C, Zhou Y, Samuelson JC. Guidelines for nucleic acid template design for optimal cell-free protein synthesis using an Escherichia coli reconstituted system or a lysate-based system. Methods Enzymol. 2021;659:351-369. doi: 10.1016/bs.mie.2021.07.005. Epub 2021 Sep 2. PMID: 34752294.</ref>. The g10 leader sequence is also present in several commercially available pET series vectors. | ||
| − | + | <br> | |
<br> | <br> | ||
[[File:T--Mingdao--2022 Registry 01.png|700px|center]] | [[File:T--Mingdao--2022 Registry 01.png|700px|center]] | ||
| − | <br> | + | <br><br> |
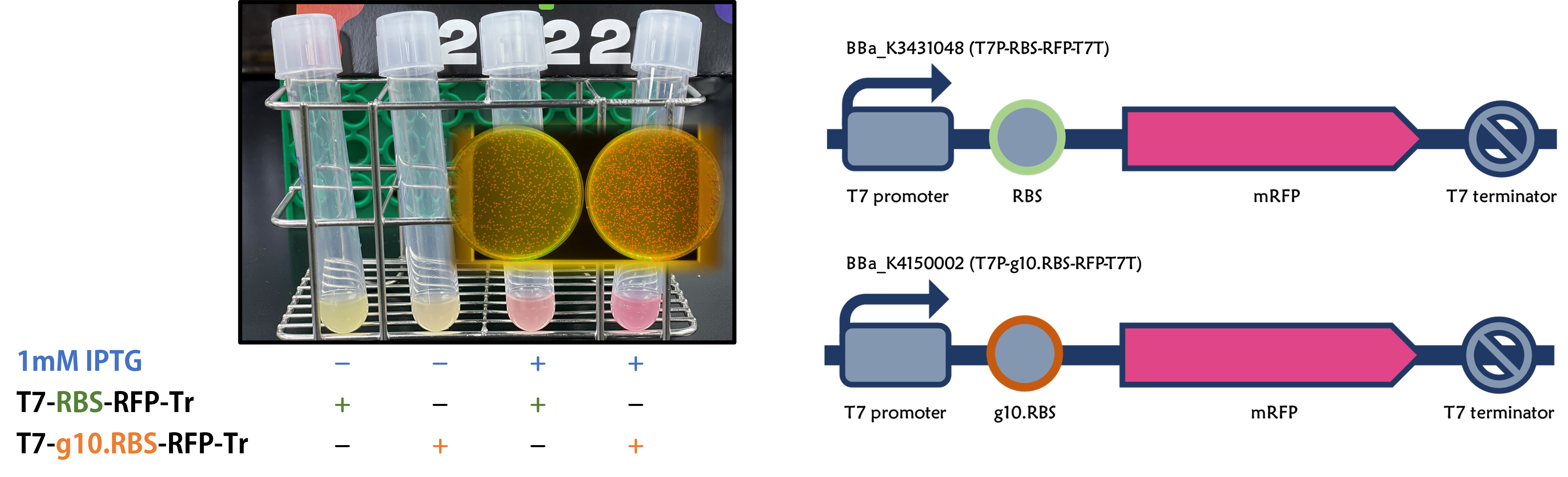
<span style="color:#00000000">This</span>We replaced the RBS with g10.RBS in BioBrick Part:BBa_K3431048 (T7-RBS-RFP-Tr) to create an improved BioBrick Part:BBa_K4150002 (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG. | <span style="color:#00000000">This</span>We replaced the RBS with g10.RBS in BioBrick Part:BBa_K3431048 (T7-RBS-RFP-Tr) to create an improved BioBrick Part:BBa_K4150002 (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG. | ||
| − | + | <br> | |
[[File:T--Mingdao--2022 Registry 02.png|950px|center]] | [[File:T--Mingdao--2022 Registry 02.png|950px|center]] | ||
| − | + | <br> | |
Figure 1. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box. | Figure 1. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box. | ||
Revision as of 01:55, 20 September 2022
T7P-g10.RBS-RFP-T7T
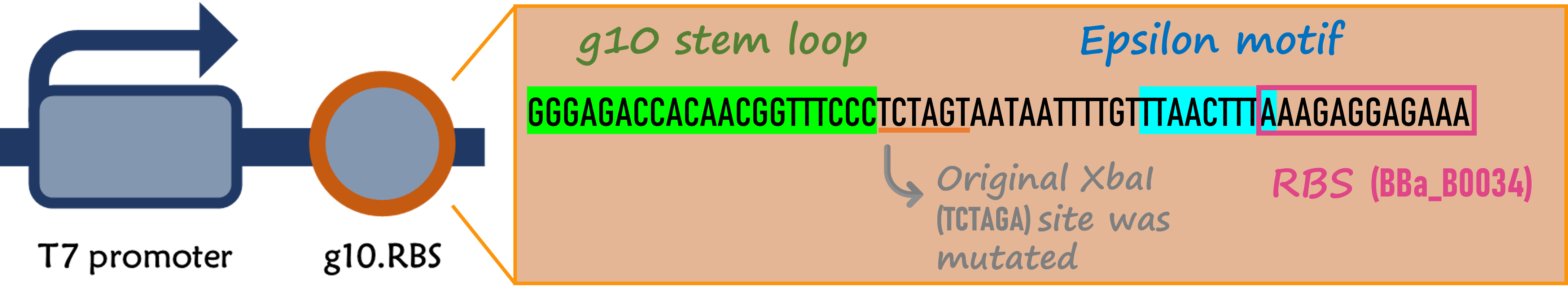
ThisThe part is a functional device composed of T7 promoter (Part:BBa_I719005), g10.RBS (Part:BBa_K4150000), mRFP (Part:BBa_E1010) and T7 terminator (Part:BBa_K731721). It's an improved version for T7 promoter + RBS + mRFP + T7 terminator (Part:BBa_K3431048)
ThisIn order to increase the expression of protein of interest, we added a canonical strong RBS (Part:BBa_B0034) with a leader sequence with a stem loop from the T7 bacteriophage gene 10 (g10.RBS, Part:BBa_K4150000).
ThisThe g10 leader sequence encodes very highly expressed phage proteins, in which 9-base of Epsilon motif exhibits perfect complementary to the 16S RNA of E. coli[1]. This structure connected with the consensus Shine-Dalgarno sequence (SD) can enhance up to 340-fold heterologous gene expression in E. coli[2]. The g10 leader sequence is also present in several commercially available pET series vectors.
ThisWe replaced the RBS with g10.RBS in BioBrick Part:BBa_K3431048 (T7-RBS-RFP-Tr) to create an improved BioBrick Part:BBa_K4150002 (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG.
Figure 1. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box.
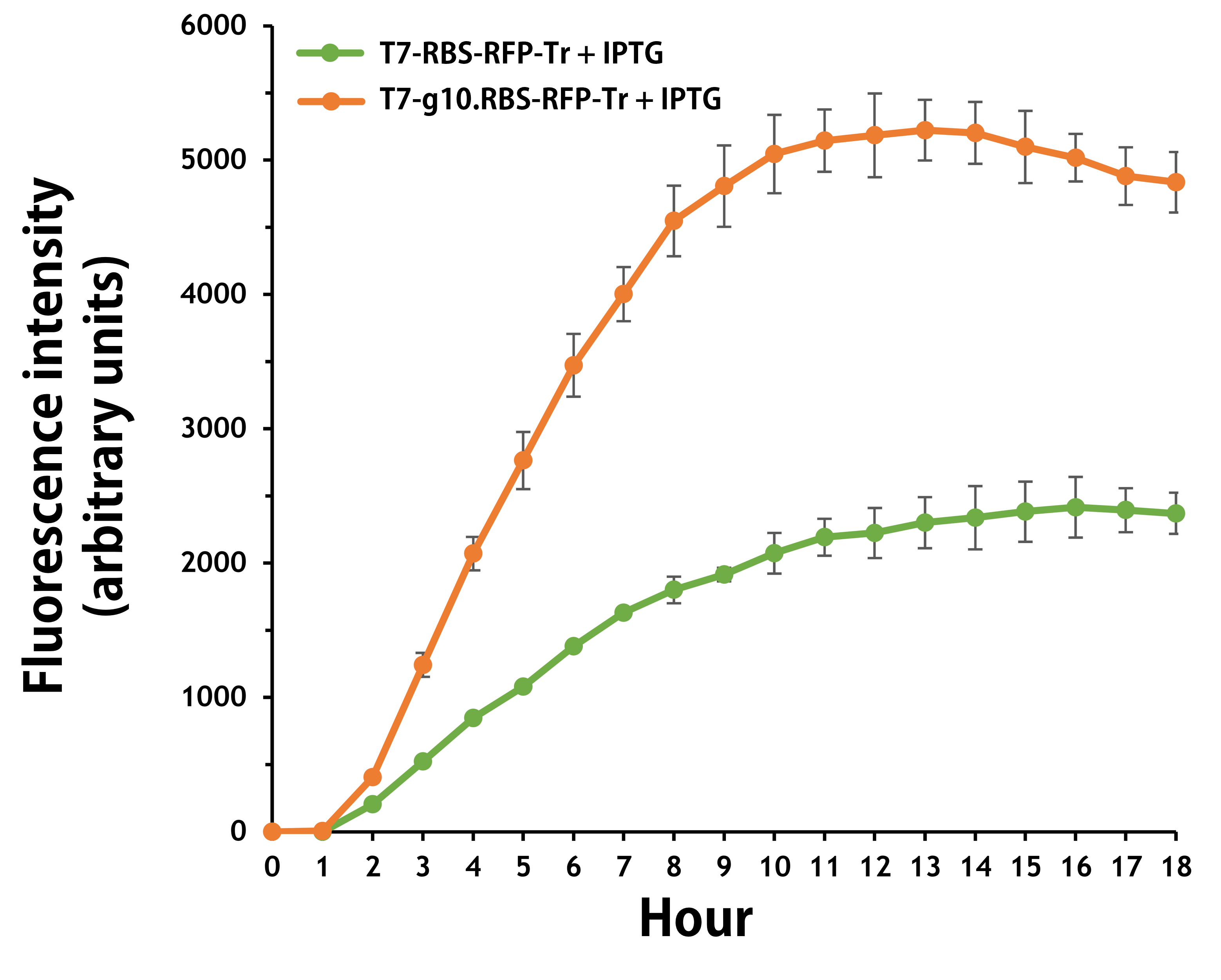
ThisFurthermore, the transformed E. coli with T7-g10.RBS-RFP-Tr compared to T7-RBS-RFP-Tr expressed RFP at faster and increased (up to 2.5-fold) fluorescence intensity levels in a time-dependent manner (Fig. 2). The above data demonstrated the g10 leader sequence in front of RBS can significantly enhance gene expression.
Figure 2. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown in LB broth supplemented with 34 μg/mL of chloramphenicol and 1mM of IPTG for 18 hours. The RFP expression levels were read at ex/em = 584/607 nm in a kinetic mode by Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments (Agilent Technologies, Inc.). The values of fluorescence intensity were presented by the data with IPTG induction minus the data without induction as background levels.
Reference
- ↑ Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988 Dec 15;73(1):227-35. doi: 10.1016/0378-1119(88)90329-0. PMID: 3072257.
- ↑ Asahara H, Magnelli P, Shi X, Tuckey C, Zhou Y, Samuelson JC. Guidelines for nucleic acid template design for optimal cell-free protein synthesis using an Escherichia coli reconstituted system or a lysate-based system. Methods Enzymol. 2021;659:351-369. doi: 10.1016/bs.mie.2021.07.005. Epub 2021 Sep 2. PMID: 34752294.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 809
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 650
Illegal AgeI site found at 762 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 34