Difference between revisions of "Part:BBa K3757011"
Erik zimmer (Talk | contribs) |
Erik zimmer (Talk | contribs) |
||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3757011 short</partinfo> | <partinfo>BBa_K3757011 short</partinfo> | ||
| − | 3in1 gene cassette encoding the cyclotide precursor protein Oak1, the cyclizing asparaginyl endopeptidase CtAEP1, and the green fluorescent protein variant sGFP (S65T). The three genes are separately regulated by a CaMV 35S promoter and 35S terminator each. CtAEP1 is equiped with a C-terminal HA-tag, Oak1 is equiped with a C-terminal c-myc tag, for affinity purification or detection in Western blot. The gene cassette is located between Agrobacterium tumefaciens left border and a right border consensus sequences. | + | 3in1 gene cassette encoding the cyclotide precursor protein Oak1, the cyclizing asparaginyl endopeptidase CtAEP1, and the green fluorescent protein variant sGFP (S65T). The three genes are separately regulated by a CaMV 35S promoter and 35S terminator each. CtAEP1 is equiped with a C-terminal HA-tag, Oak1 is equiped with a C-terminal c-myc tag, for affinity purification or detection in Western blot. The gene cassette is located between <i>Agrobacterium tumefaciens</i> left border and a right border consensus sequences. |
| − | This composite part can be used to express peptides in a cyclic form in Nicotiana benthamiana and potentially in other plants. sGFP acts as a reporter gene to monitor transfection efficiency. | + | This composite part can be used to express peptides in a cyclic form in <i>Nicotiana benthamiana</i> and potentially in other plants. sGFP acts as a reporter gene to monitor transfection efficiency. |
| Line 42: | Line 42: | ||
====Testing our System==== | ====Testing our System==== | ||
| − | The vectors were assembled by Golden Gate reactions, transformed into Escherichia coli, and plasmids were purified from overnight cultures by Miniprep. All our cloning steps were successful, as confirmed by control digest and sequencing. The results produced for these steps are shown for the example of the LIII c_blank vector in figure 1. | + | The vectors were assembled by Golden Gate reactions, transformed into <i>Escherichia coli</i>, and plasmids were purified from overnight cultures by Miniprep. All our cloning steps were successful, as confirmed by control digest and sequencing. The results produced for these steps are shown for the example of the LIII c_blank vector in figure 1. |
| − | [[Image:T--Tuebingen--results_cloning_Fig1_quer.jpeg| | + | [[Image:T--Tuebingen--results_cloning_Fig1_quer.jpeg|900px|thumb|center|'''Figure 1: Confirming the success of our Golden Gate reaction.''' (A) Colony growth. Only correct colonies are able to grow under the selective antibiotic pressure of kanamycin. If any level I vector would be present after the reaction, bacteria transformed with this vector wouldn’t be able to grow on this plate. Non-reacted level III vectors would be able to grow but can be recognized by blue-white screening, as shown in figure 2. The area in the red rectangle is shown in figure 2 in more detail. (B) Restriction digest. Digesting the by Golden Gate cloning assembled vector with HindIII leads to a different pattern compared to the empty vector before the Golden Gate reaction, as the new cyclotide insert replaces a part in the precursor that contained an additional HindIII restriction site. The restriction digest shows that all three colonies contain the insert. (C) Sequencing. To confirm the correct sequence of the synthesized insert, we sent our purified vectors of level I for sequencing. Aligning the sequences to our virtually cloned vectors in SnapGene shows that the sequence of our clone is correct. Sequencing was done with a forward (fw) and a reverse (rev) primer, red bars show correctly aligned base pairs, and white parts mismatches.]] |
| − | The correct cloning of the cyclotide with the respective AMP into our empty LIII 3in1 vector could also be verified through blue-white screening of the grown colonies on LB-agar plates supplemented with X-Gal. The empty LIII 3in1 vector contains a lac-operon, which consists of the lac promoter, lac operator, and lacZ and lacI gene, at the cloning site, where the gene sequence coding for the precursor of the cyclotide (LI constructs c_blank, c_CHEN, or c_KR-12) was inserted by Esp3I restriction enzymes. The lac promoter is inducible, therefore transcription of the genes under its control only takes place when lactose or a lactose analogon like Isopropyl-β-D-thiogalactopyranoside (IPTG) is added to the growth medium as it is otherwise repressed by a repressor encoded by the lacI gene. The lacZ gene encodes for the enzyme β-galactosidase, which hydrolyses X-Gal to galactose and a substance, that turns blue upon oxidation. 1 Hence, we could recognize successfully cloned colonies by their color because bacteria carrying the LIII 3in1 plasmid with the cyclotide precursor appeared white and not blue due to them not expressing β-galactosidase. | + | The correct cloning of the cyclotide with the respective AMP into our empty LIII 3in1 vector could also be verified through blue-white screening of the grown colonies on LB-agar plates supplemented with X-Gal. The empty LIII 3in1 vector contains a lac-operon, which consists of the lac promoter, lac operator, and lacZ and lacI gene, at the cloning site, where the gene sequence coding for the precursor of the cyclotide (LI constructs c_blank, c_CHEN, or c_KR-12) was inserted by Esp3I restriction enzymes. The lac promoter is inducible, therefore transcription of the genes under its control only takes place when lactose or a lactose analogon like Isopropyl-β-D-thiogalactopyranoside (IPTG) is added to the growth medium as it is otherwise repressed by a repressor encoded by the lacI gene. The lacZ gene encodes for the enzyme β-galactosidase, which hydrolyses X-Gal to galactose and a substance, that turns blue upon oxidation.<sup>1</sup> Hence, we could recognize successfully cloned colonies by their color because bacteria carrying the LIII 3in1 plasmid with the cyclotide precursor appeared white and not blue due to them not expressing β-galactosidase. |
[[Image:T--Tuebingen--results_cloning_Fig2.jpeg|700px|thumb|center|'''Figure 2: Blue-white screening.''' Blue-white screening enabled us to distinguish between empty level III (blue colonies) and level IV vectors (white colonies). The fact, that there are far more white than blue colonies showed us, that our Golden Gate assembly protocol is very efficient.]] | [[Image:T--Tuebingen--results_cloning_Fig2.jpeg|700px|thumb|center|'''Figure 2: Blue-white screening.''' Blue-white screening enabled us to distinguish between empty level III (blue colonies) and level IV vectors (white colonies). The fact, that there are far more white than blue colonies showed us, that our Golden Gate assembly protocol is very efficient.]] | ||
| Line 59: | Line 59: | ||
|- | |- | ||
|day 1 | |day 1 | ||
| − | |Golden Gate reaction level one (synthesised fragment into level I vector), transformation of competent E.coli cells | + | |Golden Gate reaction level one (synthesised fragment into level I vector), transformation of competent <i>E. coli</i> cells |
|- | |- | ||
|day 2 | |day 2 | ||
| Line 65: | Line 65: | ||
|- | |- | ||
|day 3 | |day 3 | ||
| − | |Miniprep, confirm correct clones by control restriction digest, Golden Gate reaction level IV, transformation of competent E.coli cells | + | |Miniprep, confirm correct clones by control restriction digest, Golden Gate reaction level IV, transformation of competent <i>E. coli</i> cells |
|- | |- | ||
|day 4 | |day 4 | ||
| Line 84: | Line 84: | ||
====Procedure==== | ====Procedure==== | ||
| − | After successful cloning, the purified vectors were transformed into Agrobacterium tumefaciens for the following agroinfiltration of tobacco plants. After growing the plants for 4 days, the recombinant protein expression of our control green fluorescent protein (GFP) was confirmed by fluorescence microscopy and the leaves were harvested for protein extraction and purification. | + | After successful cloning, the purified vectors were transformed into <i>Agrobacterium tumefaciens</i> for the following agroinfiltration of tobacco plants. After growing the plants for 4 days, the recombinant protein expression of our control green fluorescent protein (GFP) was confirmed by fluorescence microscopy and the leaves were harvested for protein extraction and purification. |
====Transformation of <i>Agrobacterium tumefaciens</i>==== | ====Transformation of <i>Agrobacterium tumefaciens</i>==== | ||
| − | The successful transformation of A. tumefaciens was confirmed by colony growth on the respective selective plates. We used the strain AGL1 which grows relatively slow, but has a high transfection rate and expression level. We successfully transformed all our level four vectors including our two negative controls for cyclisation with 2in1 vectors without the AEP gene, and 3in1 constructs containing the empty cyclotide precursor as a control (c_blank), grafted CHEN constructs (c_CHEN_1 and c_CHEN_6), and grafted KR12 constructs (c_KR-12_1 and c_KR-12_6). | + | The successful transformation of <i>A. tumefaciens</i> was confirmed by colony growth on the respective selective plates. We used the strain AGL1 which grows relatively slow, but has a high transfection rate and expression level. We successfully transformed all our level four vectors including our two negative controls for cyclisation with 2in1 vectors without the AEP gene, and 3in1 constructs containing the empty cyclotide precursor as a control (c_blank), grafted CHEN constructs (c_CHEN_1 and c_CHEN_6), and grafted KR12 constructs (c_KR-12_1 and c_KR-12_6). |
| − | [[Image:T--Tuebingen--results_ agros_3in1_16.jpeg| | + | [[Image:T--Tuebingen--results_ agros_3in1_16.jpeg|400px|thumb|center|'''Figure 1: Colony growth of <i>Agrobacterium tumefaciens</i> transformed with the 3in1 c_CHEN_6 construct on the respective selective plate.''' A bacterial suspension of <i>A. tumefaciens</i> strain AGL1 was transformed according to protocol and afterwards plated on LB agar plates containing 10 µg/ml rifampicin, 2.5 µg/ml streptomycin, 2.5 µg/ml tetracycline and 25 µg/ml kanamycin. The plates were incubated at 28 °C for two days before inoculation of an O.N. culture with the grown colonies for later infiltration of tobacco.]] |
====Infiltration of Tobacco Leaves==== | ====Infiltration of Tobacco Leaves==== | ||
| − | <i>Nicotiana benthamiana</i> leaves were infiltrated with A. tumefaciens containing the respective vectors and a p19 suppressor of gene-silencing construct. Co-transfection of p19 is essential, as p19 prevents that the addition of the vector leads to RNA-induced gene | + | <i>Nicotiana benthamiana</i> leaves were infiltrated with <i>A. tumefaciens</i> containing the respective vectors and a p19 suppressor of gene-silencing construct. Co-transfection of p19 is essential, as p19 prevents that the addition of the vector leads to RNA-induced gene silencing<sup>1</sup>. We infiltrated the tobacco leaves by syringe infiltration for transient protein expression. As a negative control, leaves were infiltrated with Agrobacteria containing only the p19 construct. |
====Fluorescence Microscopy==== | ====Fluorescence Microscopy==== | ||
| Line 101: | Line 101: | ||
After growing the plants for 4 days, the expression of our expression control protein GFP was confirmed by fluorescence microscopy. In comparison to the negative control, leaves transfected with our final vectors showed clear green fluorescence for all except the c_KR-12_6 construct (figure 2). As a negative control, non-infiltrated leaves or leaves infiltrated with p19 only were checked for green fluorescence. As shown in figure 2, this negative control showed no significant level of green fluorescence, confirming that the green fluorescence we detected for other vectors can only result from successful expression of one of our final vectors. | After growing the plants for 4 days, the expression of our expression control protein GFP was confirmed by fluorescence microscopy. In comparison to the negative control, leaves transfected with our final vectors showed clear green fluorescence for all except the c_KR-12_6 construct (figure 2). As a negative control, non-infiltrated leaves or leaves infiltrated with p19 only were checked for green fluorescence. As shown in figure 2, this negative control showed no significant level of green fluorescence, confirming that the green fluorescence we detected for other vectors can only result from successful expression of one of our final vectors. | ||
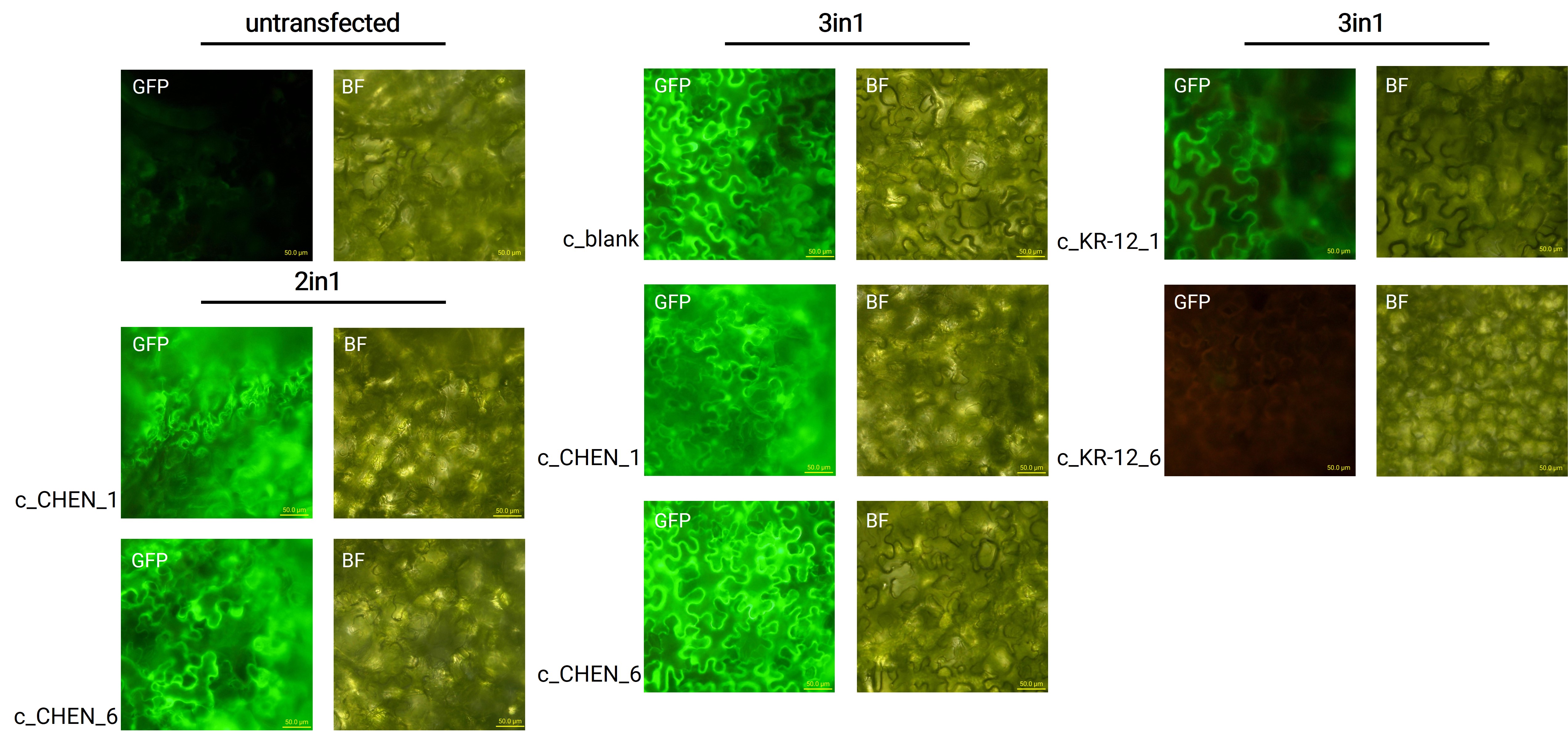
| − | [[Image:T--Tuebingen--results_expression_fluoro_figure2.jpeg|700px|thumb|center|'''Figure 2: Microscopy images from untransfected N. benthamiana leaves and leaves infiltrated with the respective 2in1 and 3in1 vectors.''' GFP – fluorescence channel, BF – brightfield microscopy, scale bar – 50.0 µm]] | + | [[Image:T--Tuebingen--results_expression_fluoro_figure2.jpeg|700px|thumb|center|'''Figure 2: Microscopy images from untransfected <i>N. benthamiana</i> leaves and leaves infiltrated with the respective 2in1 and 3in1 vectors.''' GFP – fluorescence channel, BF – brightfield microscopy, scale bar – 50.0 µm]] |
====Discussion==== | ====Discussion==== | ||
| − | We can draw the conclusion that the transfection was successful for both the 2in1 and the 3in1 vectors. As the sequences coding for our antimicrobial peptide (AMP) constructs are located on the same vector as the sequence encoding for the GFP, the AMPs should be expressed in the plant cells, as further discussed in the Extraction and Purification | + | We can draw the conclusion that the transfection was successful for both the 2in1 and the 3in1 vectors. As the sequences coding for our antimicrobial peptide (AMP) constructs are located on the same vector as the sequence encoding for the GFP, the AMPs should be expressed in the plant cells, as further discussed in the Extraction and Purification section. For the c_KR-12_6 construct, where no fluorescence was observed, infiltration was only tried once and the failure could be explained by procedural inaccuracies like insufficient resuspension of the Agrobacteria-containing infiltration medium, leading to infiltration with a too low Agrobacteria concentration. Concluding from the fluorescence microscopy results, we can summarize that our transfection strategy is efficient and works for different AMP constructs. |
However, not the whole infiltrated leaves were fluorescent but defined foci only, which indicates a low expression rate. As a further attempt to analyze the effectiveness of the agroinfiltration, GFP expression in the whole leave and also the whole plant could be analyzed under UV light. | However, not the whole infiltrated leaves were fluorescent but defined foci only, which indicates a low expression rate. As a further attempt to analyze the effectiveness of the agroinfiltration, GFP expression in the whole leave and also the whole plant could be analyzed under UV light. | ||
Furthermore, to increase the efficiency of agrotransfection and especially the expression of the desired genes, viral vectors could be used instead of a non-replicating construct for the transient expression. RNA viral-based vectors combine several advantages in one, such as the ability of the replicons to move into neighboring plant cells by cell-to-cell movement, and therefore promise high expression yields of recombinant proteins in plants. The successful Agrobacterium-mediated | Furthermore, to increase the efficiency of agrotransfection and especially the expression of the desired genes, viral vectors could be used instead of a non-replicating construct for the transient expression. RNA viral-based vectors combine several advantages in one, such as the ability of the replicons to move into neighboring plant cells by cell-to-cell movement, and therefore promise high expression yields of recombinant proteins in plants. The successful Agrobacterium-mediated | ||
| − | delivery of a tobacco mosaic virus (TMV)-based vector into N. benthamiana leaves has already been | + | delivery of a tobacco mosaic virus (TMV)-based vector into <i>N. benthamiana</i> leaves has already been shown<sup>2</sup> and it would therefore be a reasonable continuation of our project to attempt to reach higher expression yields of our cyclotides with a different vector. |
====References==== | ====References==== | ||
| Line 117: | Line 117: | ||
<sup>1</sup>Voinnet, O., Rivas, S., Mestre, P., & Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal: For Cell and Molecular Biology, 33, 949–956. https://doi.org/10.1046/j.1365-313X.2003.01676.x | <sup>1</sup>Voinnet, O., Rivas, S., Mestre, P., & Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal: For Cell and Molecular Biology, 33, 949–956. https://doi.org/10.1046/j.1365-313X.2003.01676.x | ||
| − | <sup>2</sup>Marillonnet, Sylvestre; Thoeringer, Carola; Kandzia, Romy; Klimyuk, Victor; Gleba, Yuri (2005): Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. In: Nature biotechnology 23 (6), S. 718–723. DOI: 10.1038/nbt1094. | + | <sup>2</sup>Marillonnet, Sylvestre; Thoeringer, Carola; Kandzia, Romy; Klimyuk, Victor; Gleba, Yuri (2005): Systemic <i>Agrobacterium tumefaciens</i>-mediated transfection of viral replicons for efficient transient expression in plants. In: Nature biotechnology 23 (6), S. 718–723. DOI: 10.1038/nbt1094. |
| Line 124: | Line 124: | ||
====First Attempts of Extraction and Purification of our Cyclotides==== | ====First Attempts of Extraction and Purification of our Cyclotides==== | ||
| − | The harvested leaves were frozen in liquid nitrogen and stored at - 20 °C before being pestled for extraction. The extraction was performed in HEPES buffer, whose composition was adjusted to the recommendations for the following His-purification via HisLink Protein Purification Resin (commercially available at Promega)1. Apart from the buffer we followed a protocol, which was shown to be suitable for the purification of plant | + | The harvested leaves were frozen in liquid nitrogen and stored at - 20 °C before being pestled for extraction. The extraction was performed in HEPES buffer, whose composition was adjusted to the recommendations for the following His-purification via HisLink Protein Purification Resin (commercially available at Promega)<sup>1</sup>. Apart from the buffer we followed a protocol, which was shown to be suitable for the purification of plant peptides<sup>2</sup>. |
| − | The steps of the purification were analyzed on an SDS-PAGE gel and by western blotting using an anti-His antibody. We used tricine-SDS gels that are suitable for the separation of small | + | The steps of the purification were analyzed on an SDS-PAGE gel and by western blotting using an anti-His antibody. We used tricine-SDS gels that are suitable for the separation of small peptides<sup>3</sup>. The first round of extraction and following purification was done with around 2 g of plant leaf tissue and a ratio of 1:2 plant mass:extraction buffer volume. In the extract, a total protein concentration between 0.6 to 1 mg/ml was detected. Afterwards purification via Ni-NTA was performed and the eluted fractions were then pooled and dialysed against HEPES buffer O.N. to remove the imidazole of the elution buffer. In this first experiment, no remarkable band was found in neither the Coomassie-stained SDS-PAGE gel, nor the western blot. Furthermore, the protein concentration in the eluate concentrated by centrifugation in protein concentrators (commercially available Amicon filters with a MWCO of 2 kDa) was under the detection limit of our protein quantification test and neither the crude extract nor the purified and concentrated protein showed any antimicrobial activity in our tests (data not shown here). |
[[Image:T--Tuebingen--results_coomassie_20210915.jpeg|400px|thumb|center|Figure 1: Results of SDS-PAGE with samples from the first extraction and purification. Samples from the plants infiltrated with the respective 3in1 vectors (c_blank, c_CHEN_1, c_CHEN_6) were applied to the gel either directly as crude extract or after purification via Ni-NTA resin. The flowthrough of the column, the wash step and the elution fractions as well as the pooled samples after dialysis O.N. were also analyzed. The gel was stained with Coomassie-blue to detect protein bands. M-marker, FT-flow through, CE-crude extract, W-wash, a.d.-after dialysis, E-elution, 3in1 constructs labelled according to our official nomenclature.]] | [[Image:T--Tuebingen--results_coomassie_20210915.jpeg|400px|thumb|center|Figure 1: Results of SDS-PAGE with samples from the first extraction and purification. Samples from the plants infiltrated with the respective 3in1 vectors (c_blank, c_CHEN_1, c_CHEN_6) were applied to the gel either directly as crude extract or after purification via Ni-NTA resin. The flowthrough of the column, the wash step and the elution fractions as well as the pooled samples after dialysis O.N. were also analyzed. The gel was stained with Coomassie-blue to detect protein bands. M-marker, FT-flow through, CE-crude extract, W-wash, a.d.-after dialysis, E-elution, 3in1 constructs labelled according to our official nomenclature.]] | ||
| Line 141: | Line 141: | ||
Addition of Tween20: | Addition of Tween20: | ||
| − | As already mentioned, from all the possibilities we figured out that could not have worked during our extraction and purification process, we considered the composition of our extraction buffer to be the most likely one. Therefore, we started a systematic testing process of different conditions. First, we supplemented our HEPES extraction buffer with Tween20, as this should help to extract more protein from plant | + | As already mentioned, from all the possibilities we figured out that could not have worked during our extraction and purification process, we considered the composition of our extraction buffer to be the most likely one. Therefore, we started a systematic testing process of different conditions. First, we supplemented our HEPES extraction buffer with Tween20, as this should help to extract more protein from plant tissue<sup>5</sup>. However, the addition of 0.05% Tween20 to our extraction buffer led to the same result on SDS-PAGE and western blot as before. Therefore, we used three additional buffers, namely Lämmli buffer, an acidic acetonitrile-based buffer and an adjusted SDS-buffer. |
Lämmli Buffer: | Lämmli Buffer: | ||
| Line 169: | Line 169: | ||
=====Adjusted SDS-buffer===== | =====Adjusted SDS-buffer===== | ||
| − | From the above stated results, we concluded that the extraction with a buffer supplemented with SDS seems to be the best approach. However, samples dissolved in Lämmli buffer are unsuitable for further experiments, neither purification nor assays to be able to verify the antimicrobial activity. Thus, we decided to use a slightly adjusted (only 1% SDS and no β-mercaptoethanol as reducing agent) Tris-based SDS buffer, which is recommended for the extraction of plant proteins for further use in protein quantification and | + | From the above stated results, we concluded that the extraction with a buffer supplemented with SDS seems to be the best approach. However, samples dissolved in Lämmli buffer are unsuitable for further experiments, neither purification nor assays to be able to verify the antimicrobial activity. Thus, we decided to use a slightly adjusted (only 1% SDS and no β-mercaptoethanol as reducing agent) Tris-based SDS buffer, which is recommended for the extraction of plant proteins for further use in protein quantification and analysis<sup>9</sup>. |
| − | To use the peptides extracted with this method for our antimicrobial activity assays, we needed to remove the SDS. Our first attempt was to use dialysis chambers (which we originally used to remove the imidazole after purification via Ni-NTA resin) and dialyze against a large volume of HEPES buffer O.N. as this was the buffer all previous samples and our control peptides were solubilized in | + | To use the peptides extracted with this method for our antimicrobial activity assays, we needed to remove the SDS. Our first attempt was to use dialysis chambers (which we originally used to remove the imidazole after purification via Ni-NTA resin) and dialyze against a large volume of HEPES buffer O.N. as this was the buffer all previous samples and our control peptides were solubilized in. For analysis on the SDS-PAGE we took samples prior to and after dialysis. We could show that no remarkable amount of protein was lost (figure 6). Afterwards we figured out that removal of SDS via dialysis is somehow impossible due to the interaction of the anionic detergent with cationic proteins and we followed another protocol for the removal via acetone precipitation<sup>10</sup>. Afterwards the protein pellets were resolubilized and analyzed on a gel (figure 7). We observed less bright bands here, however, this might have other reasons than loss of protein, e.g., less good staining. |
| − | [[Image:T--Tuebingen-- | + | [[Image:T--Tuebingen--results_coomassie_2021_10_06_15er.jpeg|400px|thumb|center|Figure 6: Results of SDS-PAGE with samples from extraction with adjusted SDS-buffer before and after dialysis. The leaves transfected with the respective constructs as indicated in the figure were extracted using the adjusted SDS-buffer and either applied before or after dialysis O.N. As negative controls we used plant material only transfected with control vector p19. As a positive control for the following western blot, we used a His-tagged protein. The gel was stained with Coomassie-blue after running. M-marker, p19-control vector, 3in1 constructs labelled according to our official nomenclature, His-PirC-control protein (~ 14 kDa), extr.-extraction, a.d.-after dialysis.]] |
| Line 209: | Line 209: | ||
====Conclusion==== | ====Conclusion==== | ||
| − | All in all, MALDI-TOF MS was able to confirm the presence of peptides with masses highly similar to the cyclic constructs c_CHEN_1 and c_CHEN_6, occurring in the Lämmli extract or in the acetonitrile extract, respectively. However, it is difficult to draw the conclusion that we actually detected our cyclotides, since the MS was performed with crude extracts most likely containing thousands of different peptides and proteins. We therefore presume that we may have actually detected our cyclotides using MALDI-TOF MS and western blot, but that peptide concentration is too low to see them in the LC-MS spectrum. In general, western blot is a method better suited for the detection of low abundance peptides, while mass spectrometry-based methods are used to characterize the exact mass of proteins already proven to be | + | All in all, MALDI-TOF MS was able to confirm the presence of peptides with masses highly similar to the cyclic constructs c_CHEN_1 and c_CHEN_6, occurring in the Lämmli extract or in the acetonitrile extract, respectively. However, it is difficult to draw the conclusion that we actually detected our cyclotides, since the MS was performed with crude extracts most likely containing thousands of different peptides and proteins. We therefore presume that we may have actually detected our cyclotides using MALDI-TOF MS and western blot, but that peptide concentration is too low to see them in the LC-MS spectrum. In general, western blot is a method better suited for the detection of low abundance peptides, while mass spectrometry-based methods are used to characterize the exact mass of proteins already proven to be present<sup>11</sup>. Also, the occurrence of only a single peak corresponding to the cyclotide constructs in the mass spectra fits previous MALDI-TOF results of cyclotides purified from <i>N. benthamiana</i><sup>6</sup>. |
====References==== | ====References==== | ||
Latest revision as of 02:32, 22 October 2021
3in1 Oak1 CtAEP1 sGFP (S65T)
3in1 gene cassette encoding the cyclotide precursor protein Oak1, the cyclizing asparaginyl endopeptidase CtAEP1, and the green fluorescent protein variant sGFP (S65T). The three genes are separately regulated by a CaMV 35S promoter and 35S terminator each. CtAEP1 is equiped with a C-terminal HA-tag, Oak1 is equiped with a C-terminal c-myc tag, for affinity purification or detection in Western blot. The gene cassette is located between Agrobacterium tumefaciens left border and a right border consensus sequences. This composite part can be used to express peptides in a cyclic form in Nicotiana benthamiana and potentially in other plants. sGFP acts as a reporter gene to monitor transfection efficiency.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 2239
Illegal NheI site found at 4838
Illegal NheI site found at 6434
Illegal NheI site found at 6677 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 4142
Illegal BamHI site found at 1577
Illegal BamHI site found at 1967
Illegal BamHI site found at 4398
Illegal BamHI site found at 4569 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 91
Illegal BsaI site found at 175
Illegal BsaI site found at 2326
Illegal BsaI site found at 4925
Illegal BsaI.rc site found at 6733
Contents
Usage and Biology
Our 3in1 gene cassette is designed to express cyclic peptides or proteins in plants like N. benthamiana. This is faciliated by the co-expression of three genes: The Oak1 precursor (BBa_K3669012) harbors the peptide or protein to get cyclized. A DNA-sequence encoding an arbitraty peptide can be cloned into this precursor in only one highly efficient Golden Gate cloning step. The success of cloning is thereby monitored by blue-white selection. The second gene encodes the cyclizing asparaginyl endopeptidase CtAEP1 (BBa_K3757000). This enzyme recognizes the Oak1 precursor and catalyzes cyclization of the peptide sequence embedded within it. Finally, a GFP variant sGFP (S65T) (BBa_K3757011) is co-expressed as a reporter gene. This enables an easy expression control of the three genes by fluorescence microscopy. The gene cassette is located in a binary vector between the Agrobacteria left and right border, which allow the tranfer and integration of the whole cassette into plant cells and the plant's genome, respectively. This composite part consists of the following basic and composite parts:
- A. tumefaciens LB (BBa_K233326)
- 35S Oak1 (BBa_K3757007)
- 35S CtAEP1 (BBa_K3757010)
- 35S sGFP (S65T) (BBa_K3757009)
- A. tumefaciens RB (BBa_K233327)
Cloning
Our Cloning Levels
We used four levels of Golden Gate cloning to assemble our final vectors, including green fluorescent protein (GFP), the asparagine endopeptidase (AEP), and the cyclotide precursor with the different grafted cyclotide constructs. In the first cloning level, we cloned our 35S promoter, 35S terminator, the precursor coding sequence, our antimicrobial peptide (AMP)-containing cyclotide, or the tag (HA, myc) into a LI backbone. In the second level, we assembled single genes into one vector by combining promoter, coding sequence, terminator, and tag sequences from the first level. In the third level, we now combined these whole genes into one vector. We built two kinds of level III vectors: 3in1 vectors containing the cyclotide precursor, the AEP and GFP, and 2in1 vectors containing only the precursor and GFP as a negative control. We called the vectors from this level “empty”, as they didn’t contain our AMP yet. In the last level, we now cloned our AMP into the precursor in the empty LIII vectors. We labeled these vectors according to the grafted AMP and its position. So, c_CHEN_1 indicates that the cyclotide contains the CHEN AMP in loop number 1. We called the negative control cyclotide without any grafted AMP c_blank. Furthermore, all used cyclotides contained a His-Tag in loop 5.
Why we used Golden Gate Cloning
We chose a Golden Gate based cloning method as this system not only allowed us to assemble three genes into one single vector but also to very quickly exchange the AMPs in the cyclotide precursor for grafting once the empty level III vector has been assembled. These are the most important requirements our final vector had to fulfill. We needed to express three genes at the same time: our AMP grafted into the cyclotide precursor, the AEP to convert the precursor into a cyclotide, and GFP as an expression control. In theory, these genes could also be placed on different vectors and plants could be co-infiltrated with the different systems. However, we highly preferred this 3in1 (3 genes in 1 vector) system, as this system ensures that AEP and cyclotide are expressed in the same cells, and all cells showing green fluorescence should also express the other two genes. Therefore, we regarded this system to be less error-prone and easier to handle. Building this vector by Golden Gate cloning was our preferred method, as we had access to basic vectors for this system including terminators, promoters, and tags. These vectors were kindly provided by the research group of Prof. Dr. Klaus Harter from the plant physiology at the ZMBP.
Testing our System
The vectors were assembled by Golden Gate reactions, transformed into Escherichia coli, and plasmids were purified from overnight cultures by Miniprep. All our cloning steps were successful, as confirmed by control digest and sequencing. The results produced for these steps are shown for the example of the LIII c_blank vector in figure 1.

The correct cloning of the cyclotide with the respective AMP into our empty LIII 3in1 vector could also be verified through blue-white screening of the grown colonies on LB-agar plates supplemented with X-Gal. The empty LIII 3in1 vector contains a lac-operon, which consists of the lac promoter, lac operator, and lacZ and lacI gene, at the cloning site, where the gene sequence coding for the precursor of the cyclotide (LI constructs c_blank, c_CHEN, or c_KR-12) was inserted by Esp3I restriction enzymes. The lac promoter is inducible, therefore transcription of the genes under its control only takes place when lactose or a lactose analogon like Isopropyl-β-D-thiogalactopyranoside (IPTG) is added to the growth medium as it is otherwise repressed by a repressor encoded by the lacI gene. The lacZ gene encodes for the enzyme β-galactosidase, which hydrolyses X-Gal to galactose and a substance, that turns blue upon oxidation.1 Hence, we could recognize successfully cloned colonies by their color because bacteria carrying the LIII 3in1 plasmid with the cyclotide precursor appeared white and not blue due to them not expressing β-galactosidase.
The Efficiency of our System
In the end, our established cloning system enabled us to clone different grafted AMPs in a fast and efficient way. All our cloning steps directly worked, proving the high efficiency of our protocols. After the initial cloning of the level III vectors, new AMP constructs can be cloned into the final vector within 5 days. Therefore, the new AMPs have to be synthesized, which is cost-efficient due to their small size. To clone the synthesized fragments into the level III vectors within 5 days, the following schedule should be used:
| Table | |
|---|---|
| day 1 | Golden Gate reaction level one (synthesised fragment into level I vector), transformation of competent E. coli cells |
| day 2 | Pick colonies and set overnight cultures for Miniprep for plasmid purification |
| day 3 | Miniprep, confirm correct clones by control restriction digest, Golden Gate reaction level IV, transformation of competent E. coli cells |
| day 4 | Pick colonies (blue/white screening) and set overnight cultures for Miniprep for plasmid purification |
| day 5 | Miniprep, confirm correct clones by control restriction digest -> we have our purified plasmid for Agrobacterium transformation |
References
1Julin, D. A. (2018). Blue/White Selection. In R. D. Wells, J. S. Bond, J. Klinman, & B. S. S. Masters (Eds.), Molecular Life Sciences: An Encyclopedic Reference (pp. 72–73). Springer New York. https://doi.org/10.1007/978-1-4614-1531-2_94
Expression in Nicotiana benthamiana
Procedure
After successful cloning, the purified vectors were transformed into Agrobacterium tumefaciens for the following agroinfiltration of tobacco plants. After growing the plants for 4 days, the recombinant protein expression of our control green fluorescent protein (GFP) was confirmed by fluorescence microscopy and the leaves were harvested for protein extraction and purification.
Transformation of Agrobacterium tumefaciens
The successful transformation of A. tumefaciens was confirmed by colony growth on the respective selective plates. We used the strain AGL1 which grows relatively slow, but has a high transfection rate and expression level. We successfully transformed all our level four vectors including our two negative controls for cyclisation with 2in1 vectors without the AEP gene, and 3in1 constructs containing the empty cyclotide precursor as a control (c_blank), grafted CHEN constructs (c_CHEN_1 and c_CHEN_6), and grafted KR12 constructs (c_KR-12_1 and c_KR-12_6).

Infiltration of Tobacco Leaves
Nicotiana benthamiana leaves were infiltrated with A. tumefaciens containing the respective vectors and a p19 suppressor of gene-silencing construct. Co-transfection of p19 is essential, as p19 prevents that the addition of the vector leads to RNA-induced gene silencing1. We infiltrated the tobacco leaves by syringe infiltration for transient protein expression. As a negative control, leaves were infiltrated with Agrobacteria containing only the p19 construct.
Fluorescence Microscopy
After growing the plants for 4 days, the expression of our expression control protein GFP was confirmed by fluorescence microscopy. In comparison to the negative control, leaves transfected with our final vectors showed clear green fluorescence for all except the c_KR-12_6 construct (figure 2). As a negative control, non-infiltrated leaves or leaves infiltrated with p19 only were checked for green fluorescence. As shown in figure 2, this negative control showed no significant level of green fluorescence, confirming that the green fluorescence we detected for other vectors can only result from successful expression of one of our final vectors.
Discussion
We can draw the conclusion that the transfection was successful for both the 2in1 and the 3in1 vectors. As the sequences coding for our antimicrobial peptide (AMP) constructs are located on the same vector as the sequence encoding for the GFP, the AMPs should be expressed in the plant cells, as further discussed in the Extraction and Purification section. For the c_KR-12_6 construct, where no fluorescence was observed, infiltration was only tried once and the failure could be explained by procedural inaccuracies like insufficient resuspension of the Agrobacteria-containing infiltration medium, leading to infiltration with a too low Agrobacteria concentration. Concluding from the fluorescence microscopy results, we can summarize that our transfection strategy is efficient and works for different AMP constructs.
However, not the whole infiltrated leaves were fluorescent but defined foci only, which indicates a low expression rate. As a further attempt to analyze the effectiveness of the agroinfiltration, GFP expression in the whole leave and also the whole plant could be analyzed under UV light.
Furthermore, to increase the efficiency of agrotransfection and especially the expression of the desired genes, viral vectors could be used instead of a non-replicating construct for the transient expression. RNA viral-based vectors combine several advantages in one, such as the ability of the replicons to move into neighboring plant cells by cell-to-cell movement, and therefore promise high expression yields of recombinant proteins in plants. The successful Agrobacterium-mediated delivery of a tobacco mosaic virus (TMV)-based vector into N. benthamiana leaves has already been shown2 and it would therefore be a reasonable continuation of our project to attempt to reach higher expression yields of our cyclotides with a different vector.
References
1Voinnet, O., Rivas, S., Mestre, P., & Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal: For Cell and Molecular Biology, 33, 949–956. https://doi.org/10.1046/j.1365-313X.2003.01676.x
2Marillonnet, Sylvestre; Thoeringer, Carola; Kandzia, Romy; Klimyuk, Victor; Gleba, Yuri (2005): Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. In: Nature biotechnology 23 (6), S. 718–723. DOI: 10.1038/nbt1094.
Extraction and Purification
First Attempts of Extraction and Purification of our Cyclotides
The harvested leaves were frozen in liquid nitrogen and stored at - 20 °C before being pestled for extraction. The extraction was performed in HEPES buffer, whose composition was adjusted to the recommendations for the following His-purification via HisLink Protein Purification Resin (commercially available at Promega)1. Apart from the buffer we followed a protocol, which was shown to be suitable for the purification of plant peptides2.
The steps of the purification were analyzed on an SDS-PAGE gel and by western blotting using an anti-His antibody. We used tricine-SDS gels that are suitable for the separation of small peptides3. The first round of extraction and following purification was done with around 2 g of plant leaf tissue and a ratio of 1:2 plant mass:extraction buffer volume. In the extract, a total protein concentration between 0.6 to 1 mg/ml was detected. Afterwards purification via Ni-NTA was performed and the eluted fractions were then pooled and dialysed against HEPES buffer O.N. to remove the imidazole of the elution buffer. In this first experiment, no remarkable band was found in neither the Coomassie-stained SDS-PAGE gel, nor the western blot. Furthermore, the protein concentration in the eluate concentrated by centrifugation in protein concentrators (commercially available Amicon filters with a MWCO of 2 kDa) was under the detection limit of our protein quantification test and neither the crude extract nor the purified and concentrated protein showed any antimicrobial activity in our tests (data not shown here).

Further Experiments
Scaling up
We thought that the main problem in our first purifications was the low protein concentration resulting in a too low product concentration in the concentrated eluates. Therefore, 5-6 g of leaves were used in the next extraction and the crushed leaves were incubated in the buffer for 1 h to increase extraction efficiency. The total protein concentration in the extracts was now around 1.5 mg/ml (1.44-1.71 mg/ml). To load more proteins on the SDS-PAGE, 100 µl of each sample were precipitated with acetone before loading. On the Coomassie-stained SDS-PAGE, no additional band was observed in the cyclotide-expressing plant extracts when compared to the flow through after His-purification. After purification, no protein band was visible on the gel. In the western blot, no protein band was detected in neither the crude extracts nor the purified samples.
Exchange Of Buffer Components
Addition of Tween20:
As already mentioned, from all the possibilities we figured out that could not have worked during our extraction and purification process, we considered the composition of our extraction buffer to be the most likely one. Therefore, we started a systematic testing process of different conditions. First, we supplemented our HEPES extraction buffer with Tween20, as this should help to extract more protein from plant tissue5. However, the addition of 0.05% Tween20 to our extraction buffer led to the same result on SDS-PAGE and western blot as before. Therefore, we used three additional buffers, namely Lämmli buffer, an acidic acetonitrile-based buffer and an adjusted SDS-buffer.
Lämmli Buffer:
We resuspend our in liquid nitrogen crushed plant extracts in Lämmli buffer. After a short incubation on ice, the samples were denaturated by heating to 95 °C, followed by direct application on an SDS-PAGE. All samples showed clearly more distinct bands of all kDa sizes on the Coomassie-stained SDS-gel in comparison to the prior gels, as visible in figure 2. For example, the bands in all lanes at the size of about 53 kDa are most likely from the large subunit of the enzyme Rubisco (Ribulose bisphosphate carboxylase), a plant enzyme involved in photosynthesis.

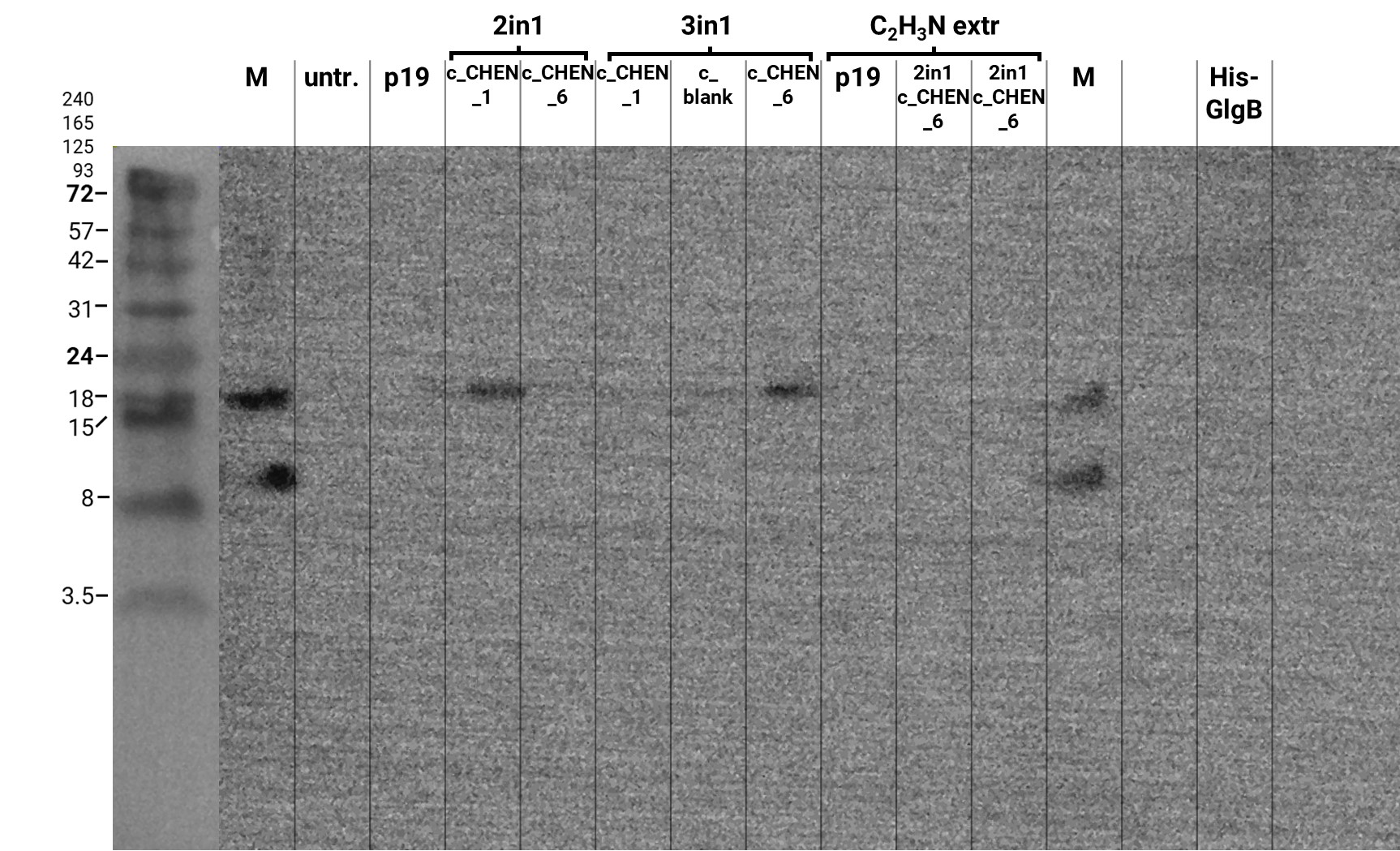
In addition, we performed a western blot of the very same samples with an anti-His-Tag primary antibody to be able to specifically detect our His-tagged cyclotides. An immunoblot signal at a size of about 18 kDa could be detected (figure 3), which raised the suspicion that cyclization of our cyclotides by the AEP did not work, as the precursor of our constructs is about the same size as the detected signal. Another possible explanation is that the band shows indeed our cyclotide, which does not run at the expected molecular range in the SDS-PAGE. This could be due to its highly positive charge, which might not be completely masked by the negatively charged SDS or due to its cyclic structure. Unfortunately, no signal was detected for our positive control, a His-tagged protein of about 84 kDa. This could be explained by the huge size difference between this control and our peptide. Thus, it could have happened that the blotting of the control on the membrane did not work.
Acidic acetonitrile-based buffer
The results from the leaves, which were extracted using an acidic-acetonitrile based buffer are also displayed on the gel and blot (Figure 2 and 3). For this method (adapted from Poon et al, 2018)6 we used dried leaf material and evaporated the extraction solvent in the last step. Prior to applying these samples onto the SDS-PAGE some of the extract was therefore solubilized in ddH2O and then mixed with 2X Lämmli buffer in equal ratio. As can be seen in figure 2, the lanes of these samples show an irregular picture, as no clear and distinct bands can be identified apart from one apparently quite big band at the height of 3.5 kDa. For these samples no signal could be detected in the western blot (figure 3).
Because of the unusual bands in the gel, we decided to repeat this experiment and applied freshly prepared samples onto a Tricine-SDS gel as well as performed a western blot afterwards. The gel looks very similar to the first attempt (compare figure 2 and 4) but on the blot we were able to detect some signal this time (figure 5). However, the molecular weight (MW) range (smaller than 3.5 kDa) in which the signal occurs does not correspond to anything we expect. Only in the lane of the construct 3in1 c_CHEN_6 a weak band can be observed at the MW of about 8 kDa, which would be around the expected size of our cyclotide construct. But as the signal is very low, this assumption would have to be verified by further experiments.
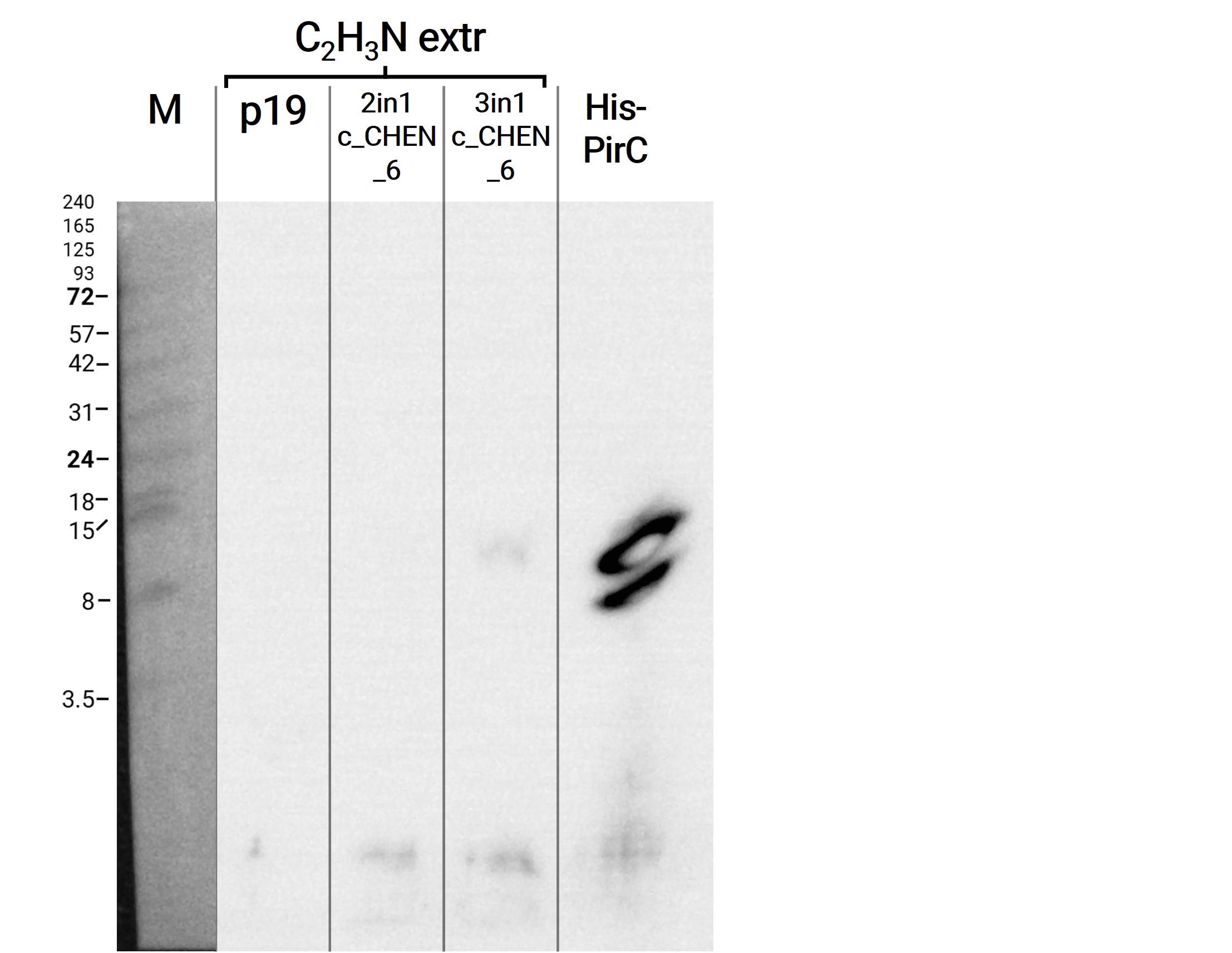
Adjusted SDS-buffer
From the above stated results, we concluded that the extraction with a buffer supplemented with SDS seems to be the best approach. However, samples dissolved in Lämmli buffer are unsuitable for further experiments, neither purification nor assays to be able to verify the antimicrobial activity. Thus, we decided to use a slightly adjusted (only 1% SDS and no β-mercaptoethanol as reducing agent) Tris-based SDS buffer, which is recommended for the extraction of plant proteins for further use in protein quantification and analysis9.
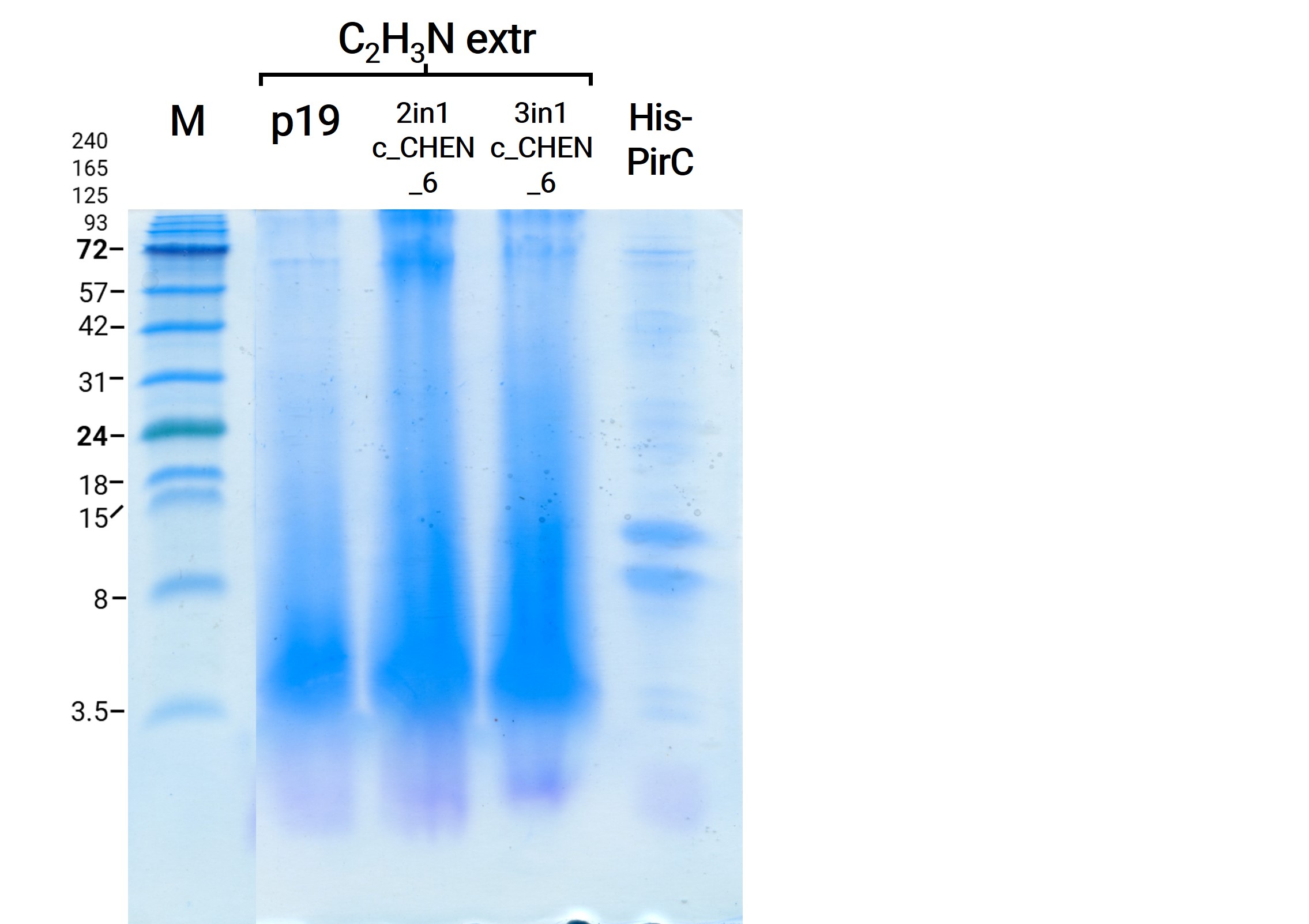
To use the peptides extracted with this method for our antimicrobial activity assays, we needed to remove the SDS. Our first attempt was to use dialysis chambers (which we originally used to remove the imidazole after purification via Ni-NTA resin) and dialyze against a large volume of HEPES buffer O.N. as this was the buffer all previous samples and our control peptides were solubilized in. For analysis on the SDS-PAGE we took samples prior to and after dialysis. We could show that no remarkable amount of protein was lost (figure 6). Afterwards we figured out that removal of SDS via dialysis is somehow impossible due to the interaction of the anionic detergent with cationic proteins and we followed another protocol for the removal via acetone precipitation10. Afterwards the protein pellets were resolubilized and analyzed on a gel (figure 7). We observed less bright bands here, however, this might have other reasons than loss of protein, e.g., less good staining.


Here we also used western blotting as a further method for analysis of the separated protein bands on the associated gels. First of all, it can be mentioned that this time we were able to see a signal of our positive control, a His-tagged protein with a size of about 14 kDa. The protein sample was unfortunately not really purified and therefore bands of other molecular weight can also be observed (figure 8). Furthermore, on the blot of the gel with the samples directly after extraction and after dialysis O.N. several bands are visible at a size of about 18 kDa and 24 kDa. As these bands are also detectable in the negative control (p19), it might be unspecific binding of the antibody. The western blot of the gel with the samples after SDS removal via precipitation showed no signal except for the positive control (data not shown).

MALDI-TOF MS
As stated above, plant crude extracts with Lämmli buffer and with acidic acetonitrile-based buffer showed protein bands with questionable identity: The 18 kDa band detected with an anti-His antibody in the Lämmli extracts, which may correspond to the cyclotide precursor or could indicate an unusual SDS-PAGE running pattern of the cyclotide itself, and the diffuse 3.5 kDa protein load visible in the Coomassie-stained acetonitrile extracts. To get more insights into the identity of these bands, mass spectrometry (MS) was performed.
Procedure
At first, gel slices at the designated height, corresponding to the protein MW marker, were cut out of the Coomassie-stained SDS-PAGE gels. For the Lämmli extracts, gel slices were cut out at the 18 kDa height, while for the acetonitrile extracts, gel slices were cut out at the 3.5 kDa height. Subsequently, the gel slices were destained, homogenized, extracted and dried from solvent in a vacuum concentrator. The dried samples were dissolved in 1:10 diluted HEPES extraction buffer, together with dried acetonitrile extracts. These samples were measured using MALDI-TOF MS.
Results
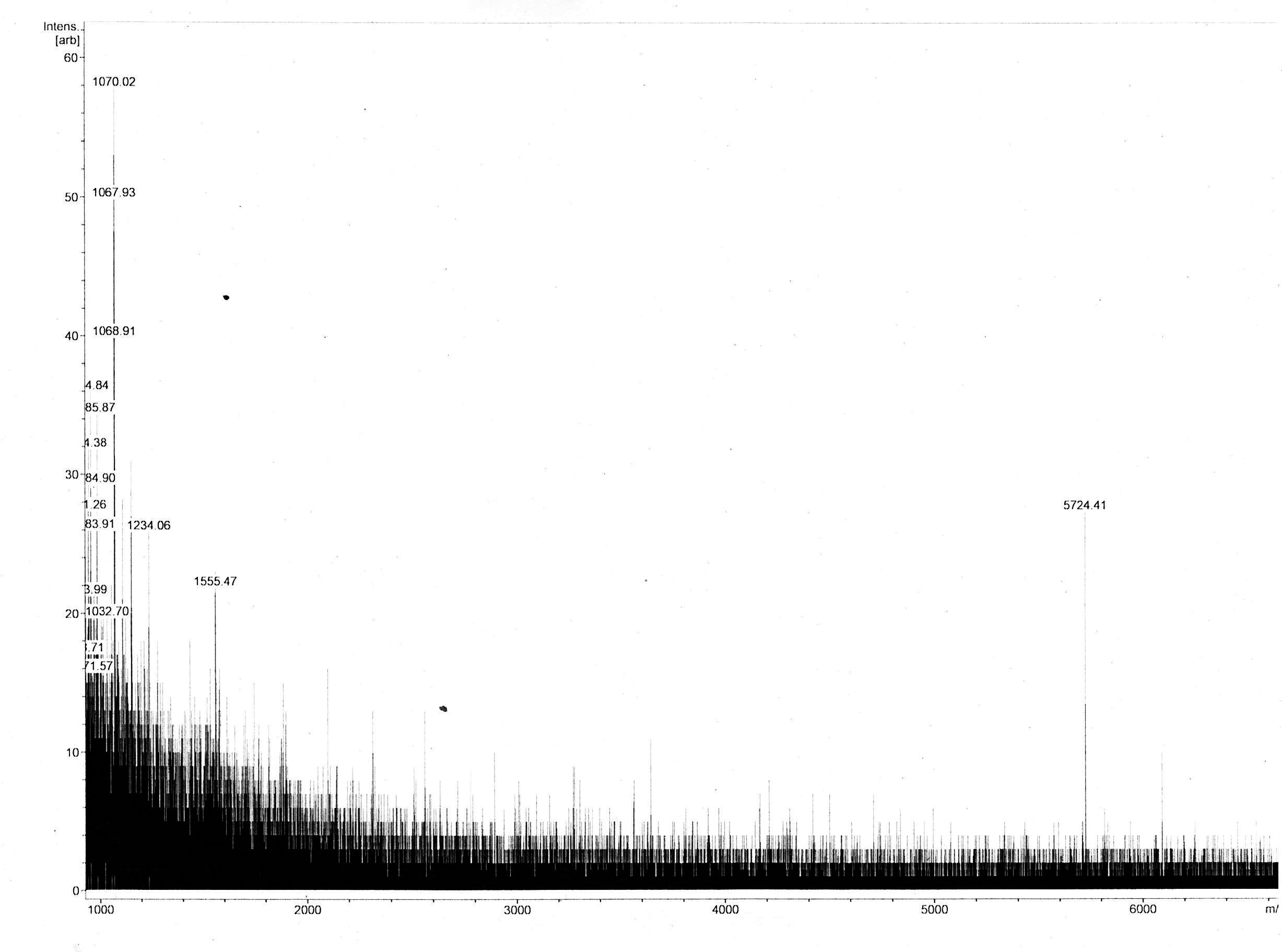
Most of the samples’ MS spectra showed no peaks with appropriate m/z ratio with relevant peak intensity. The MS spectrum of the 18 kDa Lämmli extract gel slice of 3in1 construct c_CHEN_1 showed a single peak corresponding to a m/z ratio of 5724.41 (figure 9). This value could fit with a single-charged M+ ion of cyclic c_CHEN_1, which has a theoretical average mass of 5718.30 Da with all cysteine (cys) residues reduced, as would be expected after the SDS-PAGE involving the reducing agent β-mercaptoethanol, and a theoretical average mass of 5712.30 Da with all cys residues oxidized. The deviation between theoretical mass and observed m/z ratio could be explained by the fact that the used mass spectrometer was only calibrated in the range of 2 to 4 kDa. Furthermore, it is unknown in which form our peptide occurs, since unknown modifications might be adhered. However, the running pattern in the SDS-PAGE gel does not fit the MW of our cyclotide construct. We speculated that the high positive charge of our construct, originating from its His6 tag and the cationic AMP graft, could influence its SDS running pattern.
The dissolved acetonitrile extracts were additionally concentrated and desalted using C18 resin. The MALDI-TOF MS spectrum of 2in1 construct c_CHEN_6 showed no fitting peaks. However, many peaks corresponding to m/z ratios around 3400 were visible (figure 10 A). These peaks fit single-charge M+ ions of peptides making up the broad band visible at around 3.5 kDa in the Coomassie-stained SDS-PAGE gel of the acetonitrile extracts. Therefore, we concluded that our cyclotides are unlikely to be represented by this 3.5 kDa band. The spectrum of 3in1 construct c_CHEN_6 showed a low-intensity peak with a m/z ratio of 6033.16 (figure 10 B). This value is similar to the average mass of a single-charged M+ ion of cyclic c_CHEN_6, which is expected to be 6041.89 Da with all cys residues reduced, and 6035.89 Da with all cys residues oxidized, as would be expected since no reducing agent was used on this construct.
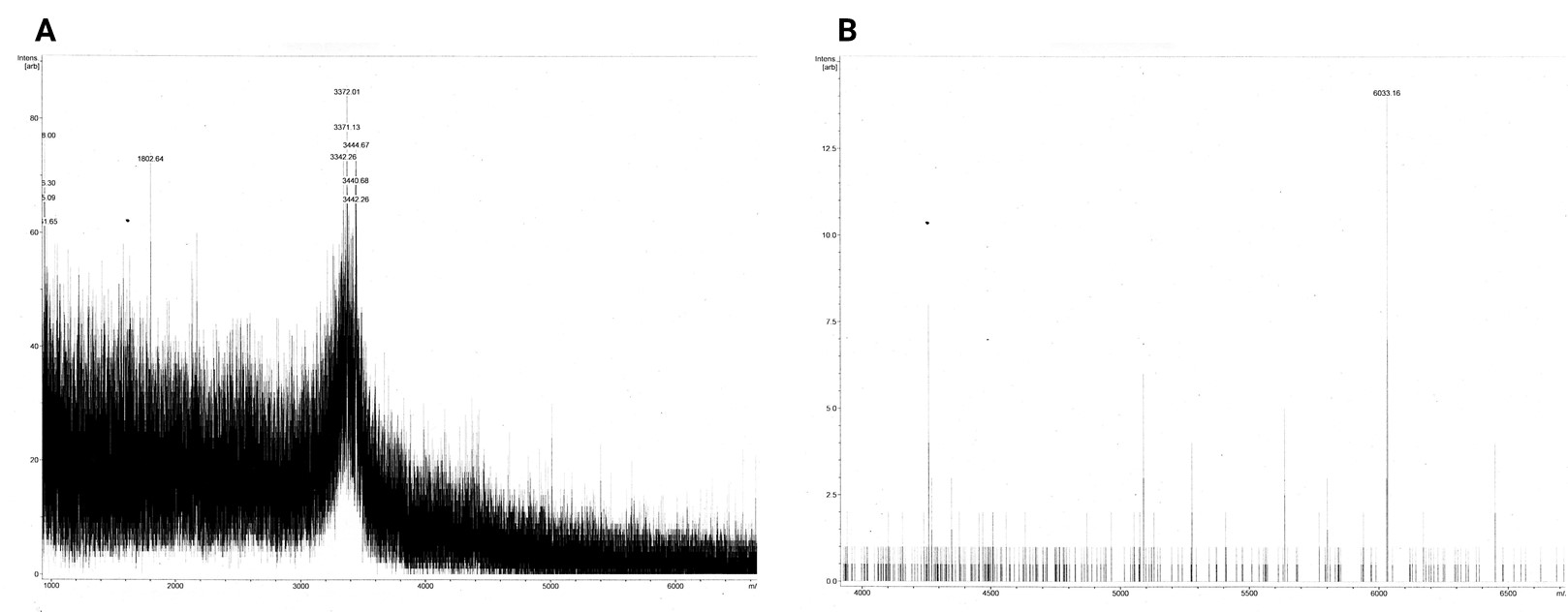
Additionally, LC-MS was performed with the concentrated and desalted acetonitrile extracts of 2in1 and 3in1 construct c_CHEN_6. However, we were not able to detect any peaks characteristic for our cyclotide constructs (data not shown).
Conclusion
All in all, MALDI-TOF MS was able to confirm the presence of peptides with masses highly similar to the cyclic constructs c_CHEN_1 and c_CHEN_6, occurring in the Lämmli extract or in the acetonitrile extract, respectively. However, it is difficult to draw the conclusion that we actually detected our cyclotides, since the MS was performed with crude extracts most likely containing thousands of different peptides and proteins. We therefore presume that we may have actually detected our cyclotides using MALDI-TOF MS and western blot, but that peptide concentration is too low to see them in the LC-MS spectrum. In general, western blot is a method better suited for the detection of low abundance peptides, while mass spectrometry-based methods are used to characterize the exact mass of proteins already proven to be present11. Also, the occurrence of only a single peak corresponding to the cyclotide constructs in the mass spectra fits previous MALDI-TOF results of cyclotides purified from N. benthamiana6.
References
1Promega Corporation: HisLink Protein Purification Resin Technical Bulletin #TB327.
2Potula, H. H. Surya Kumar; Kathuria, Sonal Roy; Ghosh, A. K.; Maiti, T. K.; Dey, S. (2008): Transient expression, purification and characterization of bioactive human fibroblast growth factor 8b in tobacco plants. In: Transgenic research 17 (1), S. 19–32. DOI: 10.1007/s11248-007-9072-4.
3Schägger, H. (2006). Tricine-SDS-PAGE. Nature Protocols, 1(1), 16–22. https://doi.org/10.1038/nprot.2006.4
4Piazza, R. M., Caetano, B. A., Henrique, C. P., Luz, D., Munhoz, D. D., Polatto, J. M., Rocha, L. B., Silva, M. A., & Mitsunari, T. (2020). Chapter 6 - Immunological tests for diarrhoea caused by diarrhoeagenic Escherichia coli targeting their main virulence factors. In C. S. Pavia & V. Gurtler (Eds.), Methods in Microbiology: Immunological Methods in Microbiology (Vol. 47, pp. 151–207). Academic Press. https://doi.org/10.1016/bs.mim.2019.11.004
5Giritch, Anatoli; Marillonnet, Sylvestre; Engler, Carola; van Eldik, Gerben; Botterman, Johan; Klimyuk, Victor; Gleba, Yuri (2006): Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. In: Proceedings of the National Academy of Sciences 103 (40), S. 14701–14706. DOI: 10.1073/pnas.0606631103.
6Poon, S., Harris, K. S., Jackson, M. A., McCorkelle, O. C., Gilding, E. K., Durek, T., van der Weerden, N. L., Craik, D. J., & Anderson, M. A. (2018). Co-expression of a cyclizing asparaginyl endopeptidase enables efficient production of cyclic peptides in planta. Journal of Experimental Botany, 69(3), 633–641. https://doi.org/10.1093/jxb/erx422
7Wiegand, Irith; Hilpert, Kai; Hancock, Robert E. W. (2008): Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. In: Nature protocols 3 (2), S. 163–175. DOI: 10.1038/nprot.2007.521.
8Bernier, Steve P.; Surette, Michael G. (2013): Concentration-dependent activity of antibiotics in natural environments. In Frontiers in microbiology 4, p. 20. DOI: 10.3389/fmicb.2013.00020.
9Conlon, H. E., & Salter, M. G. (2007). Plant Protein Extraction. In E. Rosato (Ed.), Circadian Rhythms: Methods and Protocols (pp. 379–383). Humana Press. https://doi.org/10.1007/978-1-59745-257-1_28
10Henderson, L. E., Oroszlan, S., & Konigsberg, W. (1979). A micromethod for complete removal of dodecyl sulfate from proteins by ion-pair extraction. Analytical Biochemistry, 93, 153–157. https://doi.org/10.1016/S0003-2697(79)80129-3
11Aslam, B., Basit, M., Nisar, M. A., Khurshid, M., & Rasool, M. H. (2017). Proteomics: Technologies and Their Applications. Journal of Chromatographic Science, 55(2), 182–196. https://doi.org/10.1093/chromsci/bmw167