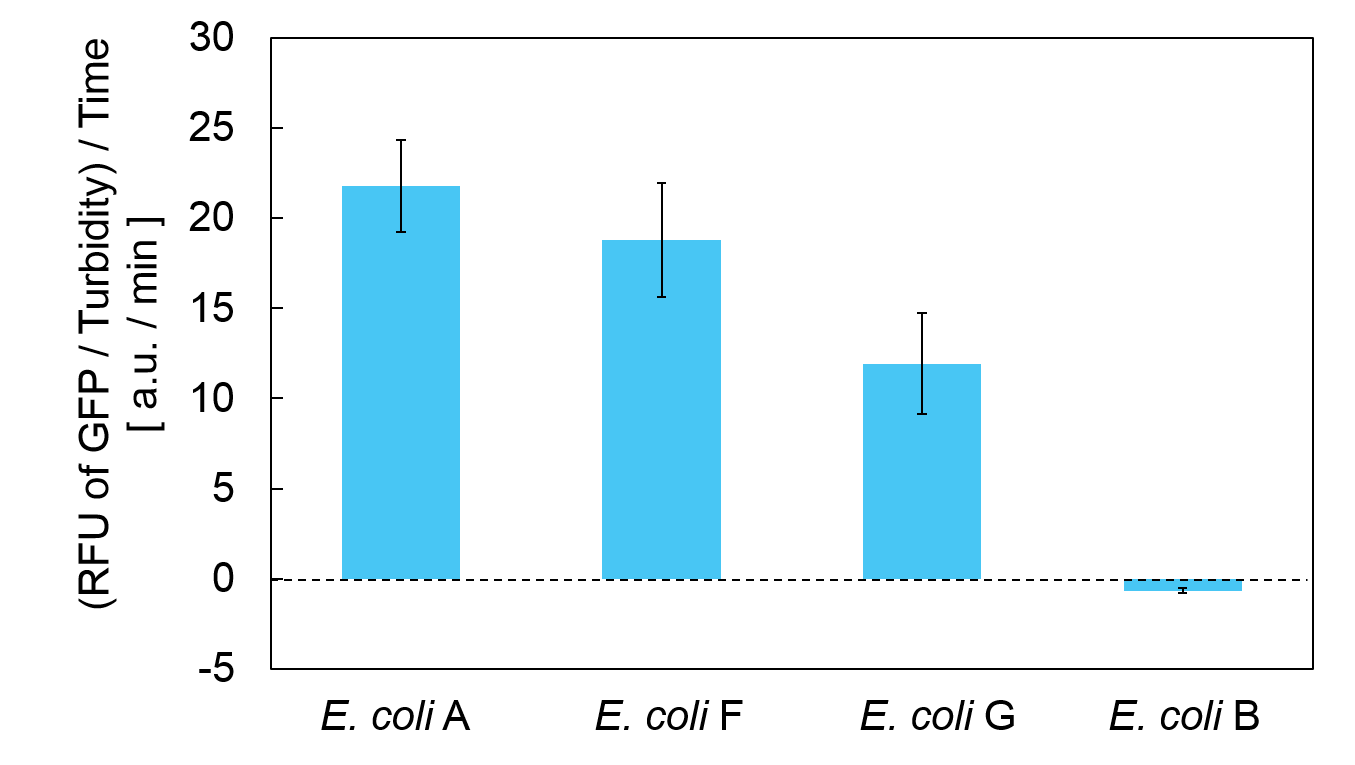Difference between revisions of "Part:BBa K1096002"
18871327112 (Talk | contribs) |
18871327112 (Talk | contribs) |
||
| Line 163: | Line 163: | ||
==Contribution From HZAU-China 2020== | ==Contribution From HZAU-China 2020== | ||
<html> | <html> | ||
| − | <body>Background | + | <body><b>Background</b> |
| − | <p>In order to fit the project, our research object for | + | <p>In order to fit the project, our research object for contribution studies is MazF protein (Escherichia coli).</p> |
| − | In our work, an iron-limiting inducible expression system was constructed. And the plasmid consists of promoter | + | <p>In our work, an iron-limiting inducible expression system was constructed. And the plasmid consists of promoter |
| − | <p>Accordingly, we aim to induce the expression of toxic gene by artificially limiting iron to achieve bacterial suicide. Finally, we successfully verified the suicide function of | + | P<sub>fhuA1</sub>(RBS and Fur box on it), toxic gene <i>mazF</i>, <i>rrnB</i> T1 terminator and T7Te terminator, which are arranged orderly on a pUC57 backbone. In this iron regulatory system, the presence of abundant iron inhibits the RNA polymerase from contacting with the promoter region sequence and prevents the transcription, so as not to express the toxic gene <i>mazF</i>. On the contrary, there is not enough iron to turn on the expression of downstream toxic gene. More importantly, this system is tightly regulated by iron-limitation signal [1].</p> |
| − | Results and analysis </P> | + | <p>Accordingly, we aim to induce the expression of toxic gene by artificially limiting iron to achieve bacterial suicide. Finally, we successfully verified the suicide function of MazF protein. |
| − | <p>First of all, we did an experiment to verify the suicide function of mazF. There were two pieces of sterile filter paper on the M9 plates, one containing 10μL | + | <p><b>Results and analysis</b> </P> |
| + | <p>First of all, we did an experiment to verify the suicide function of <i>mazF</i>. There were two pieces of sterile filter paper on the M9 plates, one containing 10μL FeSO<sub>4</sub>, and the other containing 10μL double distilled water. The M9 culture plate has not Fe<sup>2 +</sup>, so there will be bacteria growing around the filter paper (marked as the “Fe<sup>2+</sup>”) containing FeSO<sub>4</sub>, and no bacteria around the other filter paper (marked as “0”) containing double disulled water, if the plasmid has the function. Based on the experimental result (<b>Fig.1</b>), we can prove that mazF has the suicidal effect.</p> | ||
<center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF1.jpg.jpg" width="100%" alt=""> </center> | <center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF1.jpg.jpg" width="100%" alt=""> </center> | ||
| − | <p>Figure 1. E. coli growth with toxic gene mazF on the M9 medium plates in the filter paper diffusion experiment. The number of colony forming unit around the two filter paper, one containing 10 uL FeSO4 and another containing 10 uL double distilled water. This experiment was repeated for three times.</p> | + | <p><b>Figure 1. E. coli growth with toxic gene mazF on the M9 medium plates in the filter paper diffusion experiment.</b> The number of colony forming unit around the two filter paper, one containing 10 uL FeSO4 and another containing 10 uL double distilled water. This experiment was repeated for three times.</p> |
| − | <p>From figure 1, we can clearly see that there are more bacteria growing around the filter paper, marked as | + | <p>From <b>figure 1</b>, we can clearly see that there are more bacteria growing around the filter paper, marked as “Fe<sup>2+</sup>”, while there is no bacteria around the filter paper marked as “0”. It can be concluded that MazF protein is effective under the control of the Fe<sup>2+</sup>.</p> |
| − | <p>In addition, we set up four groups of different concentrations of | + | <p>In addition, we set up four groups of different concentrations of Fe<sup>2 +</sup> to regulate the expression of <i>mazF</i> to further prove the suicide effect. If mazF has the suicidal effect, the growth of bacteria will be inhibited in the absence of Fe<sup>2 + </sup>(0 μM Fe<sup>2+</sup>). According to the experimental results (,<i>Fig. 2</i>), the turbidity degree of bacteria has obvious difference under different conditions of Fe<sup>2+</sup>. Compared with the control, the turbidity of “0 μM Fe<sup>2+</sup>”group was a little clearer than that in the “10 μM, 20 μM, 40 μM Fe<sup>2+</sup>” groups.</p> |
<center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF2.jpg.jpg" width="100%" alt=""> </center> | <center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF2.jpg.jpg" width="100%" alt=""> </center> | ||
| − | <p>Figure 2. E. coli growth with toxic gene mazF in the M9 medium with the different concentration gradients | + | <p><b>Figure 2. E. coli growth with toxic gene <i>mazF</i> in the M9 medium with the different concentration gradients FeSO<sub>4</sub> Turbidity degree of bacterial liquid in each group. This experiment was repeated for three times.</b></p> |
| − | <p>Worried that the addition of | + | <p>Worried that the addition of Fe<sup>2+</sup> will affect the growth of common bacteria, so we also conducted a control test of DH5α(<i>Fig. 3</i>). Within 9 hours of the incubating, OD<sub>600</sub> of each group was measured every hour. From <i>Fig.3</i>, the growth of the “0 μM Fe<sup>2+</sup>” group is inhibited in the logarithmic period. And the growth of DH5α in the 4 groups of Fe<sup>2+</sup> were showing on no difference.</p> |
<center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF3.jpg.jpg" width="100%" alt=""> </center> | <center><img src="https://2020.igem.org/wiki/images/c/c6/T--HZAU-China--MazF3.jpg.jpg" width="100%" alt=""> </center> | ||
| − | <p>Figure 3. Growth curve of bacteria with different concentrations of Fe2+. A. Growth curve of DH5α. At 0 h, different concentration gradients | + | <p><b>Figure 3. Growth curve of bacteria with different concentrations of Fe2+.</b> A. Growth curve of DH5α. At 0 h, different concentration gradients FeSO<sub>4</sub> were added. The error bars represent the standard deviation (SD) for three independent experiments, performed in triplicate. B. E. coli growth with toxic gene mazF. Growth curves to demonstrate the tightly controllable expression of toxic gene ,<i>mazF</i> under repression (+Fe,10μM , 20 μM, 40 μM FeSO<sub>4</sub>), or induction (0 μM FeSO<sub>4</sub>) . At 0 h, the different concentration gradients FeSO<sub>4</sub> were added.</p> |
| − | <p>In conclusion, the expression of toxic gene mazF is related to the concentration of | + | <p>In conclusion, the expression of toxic gene <i>mazF</i> is related to the concentration of Fe<sup>2 +</sup>.And when there is enough Fe<sup>2+ </sup>in the culture environment, the expression is repressed. On the contray, when the amount of Fe<sup>2 +</sup> is not enough, the gene expression is induced so as to express toxic protein MazF to kill the bacteria. </p> |
| − | <p>Methods</p> | + | <p><b>Methods</b></p> |
| − | <p>Plasmids Construction</p> | + | <p><b>Plasmids Construction</b></p> |
| − | <p>The promoter | + | <p>The promoter P<sub>fhuA1</sub> (RBS and Fur box on it) and <i>mazF</i> are ligated between the parts by overlap extension PCR, and the construted fragment is ligated to pUC57 by homologous recombination. The correct construction is confirmed by sequencing.</p> |
| − | <p>Inoculation and cultivation</p> | + | <p><b>Inoculation and cultivation</b></p> |
<p>1. Methods of molecular cloning and transformation are described above. Transform this plasmid into E. coli DH5α. Then spread it onto a LB medium plates with 60 μg/mL ampicillin and incubate overnight at 37℃ in a biochemical incubator.</p> | <p>1. Methods of molecular cloning and transformation are described above. Transform this plasmid into E. coli DH5α. Then spread it onto a LB medium plates with 60 μg/mL ampicillin and incubate overnight at 37℃ in a biochemical incubator.</p> | ||
| − | <p>2. Pick three monoclonals from the same plate as parallel repeats. With the 5 ml LB medium containing 60 μg/mL ampicillin, the seed culture media are grow at 37℃ while shaking at | + | <p>2. Pick three monoclonals from the same plate as parallel repeats. With the 5 ml LB medium containing 60 μg/mL ampicillin, the seed culture media are grow at 37℃ while shaking at 200 rpm overnight.</p> |
<p>3. Take 2 mL seed culture medium into 2 mL EP tube and centrifuge for 2 minutes at 5000 rpm. Then wash the centrifugated deposit with PBS buffer to remove ferrous ions for three times. After that resuspend the deposit in the M9 medium (60 μg/ml ampicillin) and make the final volume 5 mL.</p> | <p>3. Take 2 mL seed culture medium into 2 mL EP tube and centrifuge for 2 minutes at 5000 rpm. Then wash the centrifugated deposit with PBS buffer to remove ferrous ions for three times. After that resuspend the deposit in the M9 medium (60 μg/ml ampicillin) and make the final volume 5 mL.</p> | ||
| − | <p>4. Measure the | + | <p>4. Measure the OD<sub>600</sub> value of the resuspending culture media in an automatic microplate reader (SynergyH1 hybrid multimodal reader). Then dilute the resuspending culture medium to OD<sub>600</sub> = 0.02 and add FeSO<sub>4</sub> solution (40 mM) to the final concentration of 0 μM, 10 μM , 20 μM, 40 μM. The blank control groups are the same volume of M9 medium containing 60 μg/mL ampicillin which are added the corresponding concentration gradient FeSO<sub>4</sub> solution respectively.</p> |
| − | <p>5. Incubate the cultures for 9 h at 37℃ while shaking at | + | <p>5. Incubate the cultures for 9 h at 37℃ while shaking at 200 rpm.</p> |
| − | <p>6. Samples are taken from each group once an hour and measured by UV spectrophotometer. And then convert the raw data to | + | <p>6. Samples are taken from each group once an hour and measured by UV spectrophotometer. And then convert the raw data to OD<sub>600</sub>.</p> |
| − | <p>7. Take 50uL from the seed culture medium and add it into 5ml LB medium containing 60 μg/mL ampicillin. Incubate for 8-9 hours at 37℃ while shaking at | + | <p>7. Take 50uL from the seed culture medium and add it into 5ml LB medium containing 60 μg/mL ampicillin. Incubate for 8-9 hours at 37℃ while shaking at 200 rpm.</p> |
| − | <p>8. Dilute the incubated bacterial fluid to | + | <p>8. Dilute the incubated bacterial fluid to 10<sup>2</sup> and 10<sup>3</sup> times respectively.</p> |
| − | <p>9. Take 10 uL bacterial fluid from the two diluted groups, spread it onto M9 medium containing 60 μg/mL ampicillin respectively. Then two filter papers with a drop of 10 uL of | + | <p>9. Take 10 uL bacterial fluid from the two diluted groups, spread it onto M9 medium containing 60 μg/mL ampicillin respectively. Then two filter papers with a drop of 10 uL of FeSO<sub>4</sub> (40 mM) and 10 uL of double distilled water respectively are put onto each plate. Blank controls are fresh M9 medium containing ampicillin (Amp, 60 μg/mL) with the engineered bacteria. Incubate them overnight at 37℃ in a biochemical incubator. </p> |
<p>10. All experiments above are carried out in 3 biological replications.</p> | <p>10. All experiments above are carried out in 3 biological replications.</p> | ||
Revision as of 01:28, 28 October 2020
MazF protein (E. coli)
MazF is toxin protein in MazE-MazF, toxin-antitoxin module. It is function as mRNA endonuclease
Characterization
Group: Tokyo Tech 2016
Author: Kazuki Fujisawa
We designed parts(BBa_K1949100, BBa_K1949101, BBa_K1949102, BBa_K1949103, BBa_K1949104) to characterize mazE(BBa_K1096001) and mazF(BBa_K1096002).
Characterize
We performed experiment to work our final genetic circuits and characterized mazE and mazF.
This experiment consists of the four parts below.
Ⅰ.Adjustment of mazF Expression
Ⅱ.mazEF System Assay ~Stop & GO~
Ⅲ.mazEF System Assay ~Go & Stop~
Ⅳ.mazEF System Assay on the LB Agar Plate (Queen's Caprice)
Ⅰ.Adjustment of mazF Expression
In order to control cell growth as we desire using the mazEF system, it is necessary to adjust the expression level of mazF. It has been reported that MazF has very strong ability to inhibit cell growth and that mazE expression can not recover it when mazF is expressed at high level[1]. Therefore, we here explored the relationship between concentration of the expression inducer for mazF (arabinose in this experiment) and expression level of it; such information is important for operating our final genetic circuits properly.
We constructed
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
It was found that cell growth of E. coli B was inhibited under arabinose (inducer for mazF) concentrations of over 0.02% (Fig. 1). Interestingly, at arabinose concentration of 0.2%, GFP fluorescence intensity fell markedly, regardless of mazF expression, suggesting that high arabinose concentrations may inhibit gfp expression or prevent GFP from exerting fluorescence. Taken together, it is concluded that the concentration of arabinose should be 0.02%.
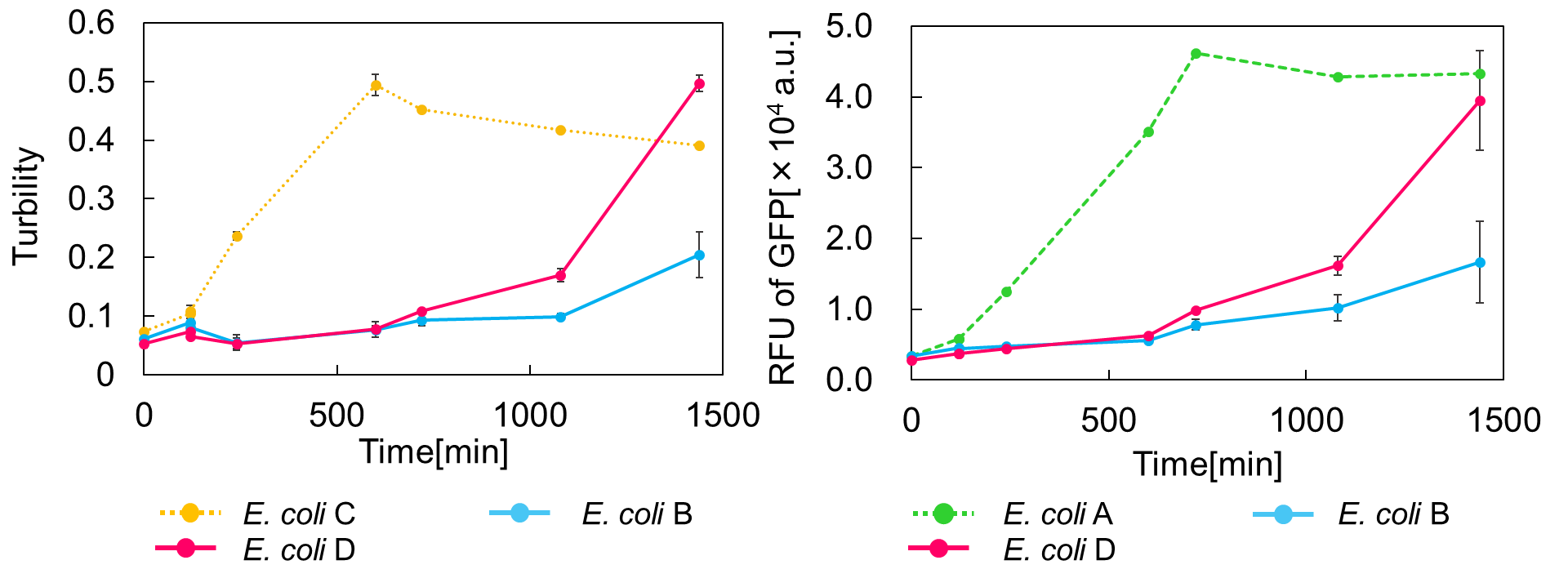
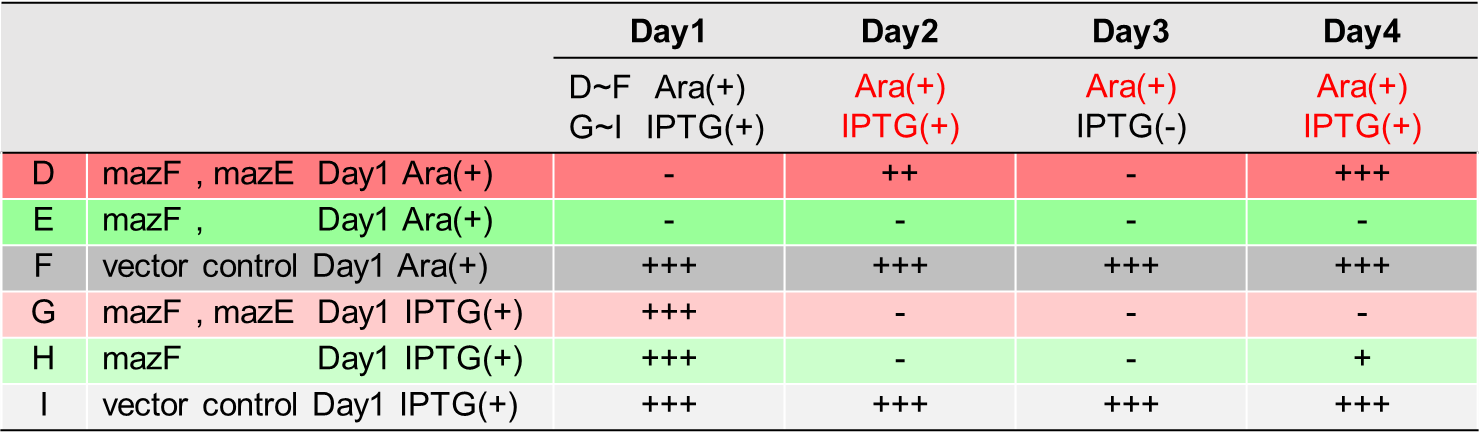
Ⅱ.mazEF System Assay ~Stop & GO~
The most attractive feature of the mazEF system is that cytotoxity of a toxin protein (MazF) is determined by the stoichiometric ratio of a toxin protein to a corresponding antitoxin protein (MazE) in cells. The story of Snow White involves sleep and resuscitate from sleep, and thus, we think that this feature is useful for our project.
We constructed
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli D: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs - mazE (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
As shown in Fig. 2, when the mazE expression was induced 2 h after the mazF expression, cell growth resumed gradually. Similarly, the RFU of GFP also increased mazF expression, and about 8 h later, cell growth resumed. Similarly, the RFU of GFP was also resumed. Before starting the experiments, we anticipated that recovery from the growth inhibition by mazF would be observed immediately after the induction of mazE expression. However, it took 8 h for us to observe the recovery. The reason is unclear, but the prior expression of mazF might damage the cells and it might take time to repair. Alternatively, the mazF expression might lead cells to the dormant state from which cells can hardly escape. We assume that this feature is advantageous for our Snow White project, because we can prolong the sleeping time of Snow White and take sufficient time for Prince to find Snow White. As a conclusion, the “Stop and Go” experiment was successful.
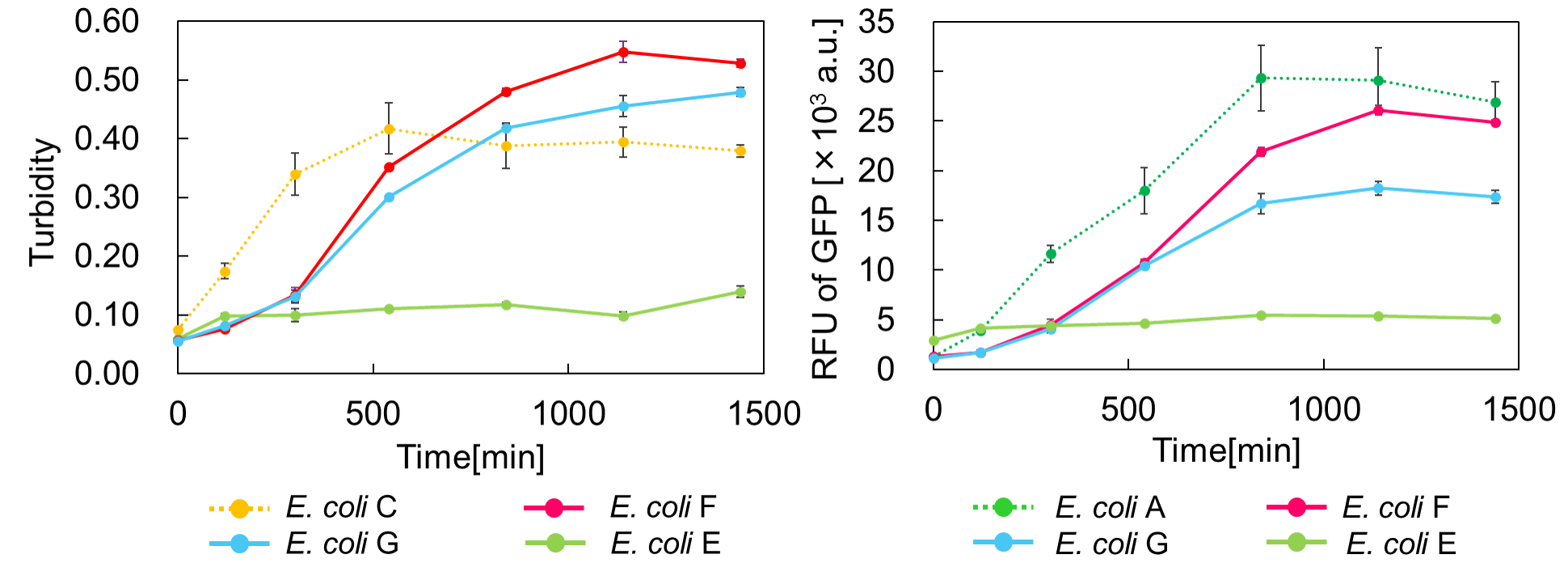
Ⅲ.mazEF System Assay ~Go & Stop~
In experimentⅡ, a toxin inhibits cell growth, and an antitoxin resuscitates it. However, what will happen when a toxin is expressed after the constitutive expression of antitoxin? Therefore, in the experiments of this section, we conducted a reciprocal experiment and named this experiment "Go & Stop".
We constructed
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli G: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs(weak) - mazE (pSB3K3)
E. coli F: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs - mazE (pSB3K3)
E. coli E: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), vector (pSB3K3).
result
Turbidity of E. coli G stopped earlier than that of E. coli F (Fig. 3), indicating that cellular mazE amount over MazF amount affects cytotoxity of MazF. In other words, the cytotoxity of MazF is determined stoichiometrically. Similarly, final RFU of E. coli G was lower than that of E. coli F. From the results of ExperimentⅠ and ExperimentⅡ, it is expected that mazEF can repeatedly control cell growth.
Calculation of the change of RFU of GFP / Turbidity per unit time (translation efficiency) indicates that the expression level of mazE correlated with the translation efficiency(Fig. 4).
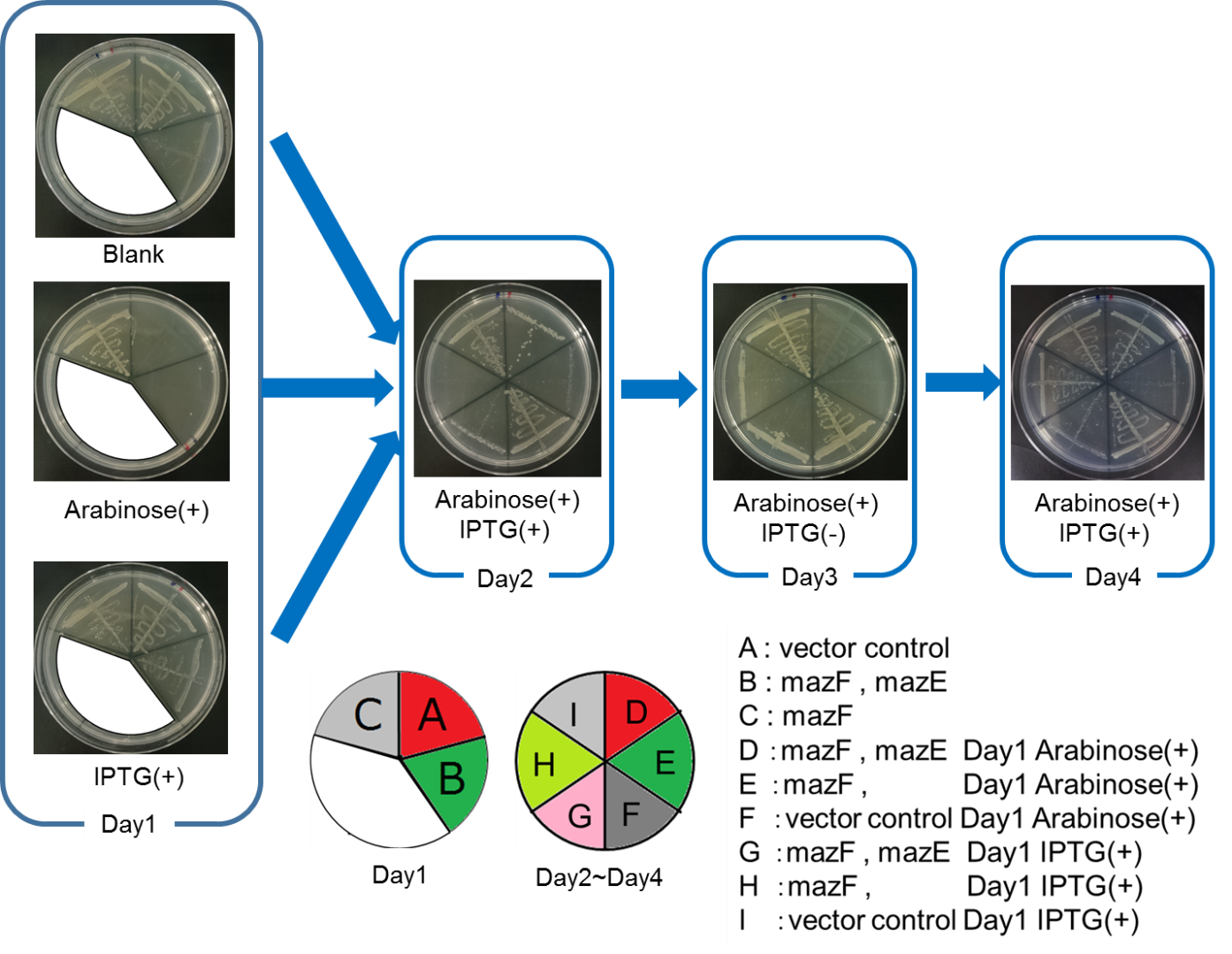
Ⅳ.mazEF System Assay on the LB Agar Plate(Queen's Caprice)
The control of cell growth by the mazEF system has been shown until the previous sections. In this section, we analyzed whether the “stop & go” experiment can be repeated many times. It seemed like that dormancy of Snow White was controled by Queen, we named this experiment "Queen's caprice".
We constructed
E. coli i: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli D: PBAD - rbs - mazF (pSB6A1), Plac - rbs - mazE (pSB3K3)
E. coli H: PBAD - rbs - mazF(pSB6A1), Plac - rbs (pSB3K3).
result
From the result, it was clarified that growth of E. coli cells was repeatedly controlled by expression of mazE (Fig. 5 and Fig. 6). We believe that the repeated control of cell growth is very useful for the future biotechnology; see http://2016.igem.org/Team:Tokyo_Tech/Human_Practices.
reference
[1] Hazan, R., B. Sat, and H. Engelberg-Kulka. E. coli mazEF mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004 Dec;186(24):8295-8300.
Contribution From XMU-China 2020
Group: iGEM Team XMU-China 2020
Author: Shi Zhang
Summary: colony forming unit (CFU)
Characterization from iGEM20-XMU-China
MazF is toxin protein in MazF-MazE, toxin-antitoxin module. It was first registered in 2013 and used as an mRNA endonuclease. It kills bacteria without cracking them, which will directly influence the value of OD600.
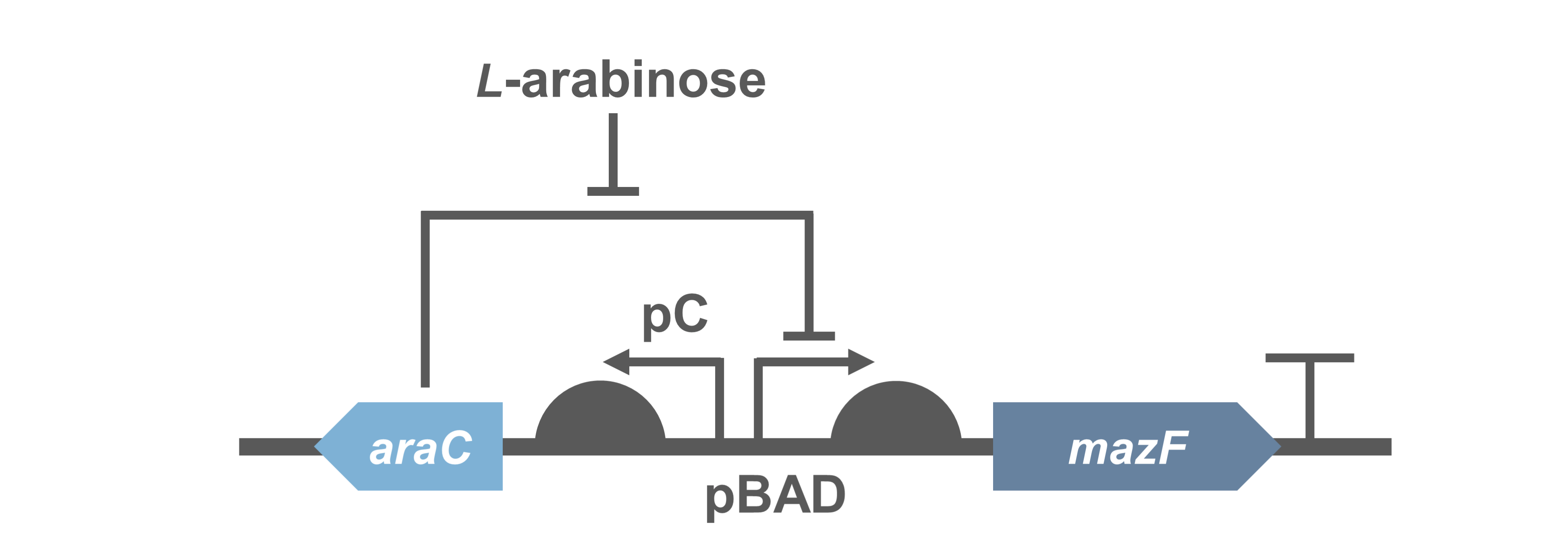
In order to quantify the toxicity of mazF, colony forming unit (CFU) cell viability assays were used to measure functionality of the circuit(1).
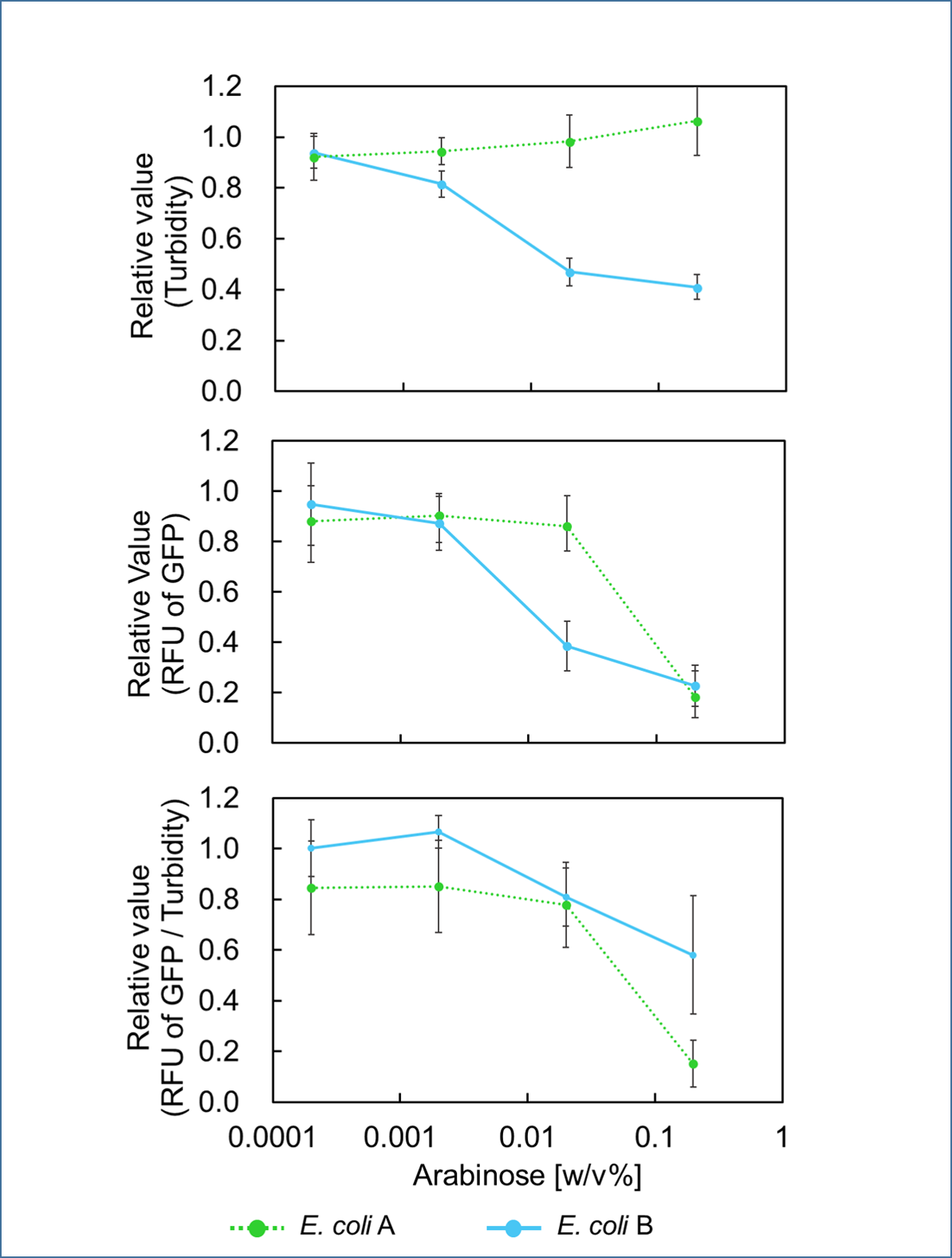
Because direct expression of MazF will kill bacteria, we construct the circuit "PBad/araC-RBS-MazF-terminator-pSB1C3"(BBa_K3332083) and "PBad/araC-RBS-EYFP-terminator-pSB1C3"(BBa_K3332082) in E.coli BL21(DE3) to characterize its function.
- Fig.1 genetic circuit of PBad/araC-RBS-MazF-terminator.
When 0.2% arabinose was added, the experimental group showed a sharp decrease on the CFU, which indicates the protein encoded by MazF is fatal to E. coli. It further demonstrated that the MazF encoded by J23100-RBS (BBa_K880005) was able to work as a toxin.
- Fig.2 The results of CFU.The round dot indicates the non induction group, and the square dot indicates the induction group.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contribution From HZAU-China 2020
Background
In order to fit the project, our research object for contribution studies is MazF protein (Escherichia coli).
In our work, an iron-limiting inducible expression system was constructed. And the plasmid consists of promoter PfhuA1(RBS and Fur box on it), toxic gene mazF, rrnB T1 terminator and T7Te terminator, which are arranged orderly on a pUC57 backbone. In this iron regulatory system, the presence of abundant iron inhibits the RNA polymerase from contacting with the promoter region sequence and prevents the transcription, so as not to express the toxic gene mazF. On the contrary, there is not enough iron to turn on the expression of downstream toxic gene. More importantly, this system is tightly regulated by iron-limitation signal [1].
Accordingly, we aim to induce the expression of toxic gene by artificially limiting iron to achieve bacterial suicide. Finally, we successfully verified the suicide function of MazF protein.
Results and analysis
First of all, we did an experiment to verify the suicide function of mazF. There were two pieces of sterile filter paper on the M9 plates, one containing 10μL FeSO4, and the other containing 10μL double distilled water. The M9 culture plate has not Fe2 +, so there will be bacteria growing around the filter paper (marked as the “Fe2+”) containing FeSO4, and no bacteria around the other filter paper (marked as “0”) containing double disulled water, if the plasmid has the function. Based on the experimental result (Fig.1), we can prove that mazF has the suicidal effect.

Figure 1. E. coli growth with toxic gene mazF on the M9 medium plates in the filter paper diffusion experiment. The number of colony forming unit around the two filter paper, one containing 10 uL FeSO4 and another containing 10 uL double distilled water. This experiment was repeated for three times.
From figure 1, we can clearly see that there are more bacteria growing around the filter paper, marked as “Fe2+”, while there is no bacteria around the filter paper marked as “0”. It can be concluded that MazF protein is effective under the control of the Fe2+.
In addition, we set up four groups of different concentrations of Fe2 + to regulate the expression of mazF to further prove the suicide effect. If mazF has the suicidal effect, the growth of bacteria will be inhibited in the absence of Fe2 + (0 μM Fe2+). According to the experimental results (,Fig. 2), the turbidity degree of bacteria has obvious difference under different conditions of Fe2+. Compared with the control, the turbidity of “0 μM Fe2+”group was a little clearer than that in the “10 μM, 20 μM, 40 μM Fe2+” groups.

Figure 2. E. coli growth with toxic gene mazF in the M9 medium with the different concentration gradients FeSO4 Turbidity degree of bacterial liquid in each group. This experiment was repeated for three times.
Worried that the addition of Fe2+ will affect the growth of common bacteria, so we also conducted a control test of DH5α(Fig. 3). Within 9 hours of the incubating, OD600 of each group was measured every hour. From Fig.3, the growth of the “0 μM Fe2+” group is inhibited in the logarithmic period. And the growth of DH5α in the 4 groups of Fe2+ were showing on no difference.

Figure 3. Growth curve of bacteria with different concentrations of Fe2+. A. Growth curve of DH5α. At 0 h, different concentration gradients FeSO4 were added. The error bars represent the standard deviation (SD) for three independent experiments, performed in triplicate. B. E. coli growth with toxic gene mazF. Growth curves to demonstrate the tightly controllable expression of toxic gene ,mazF under repression (+Fe,10μM , 20 μM, 40 μM FeSO4), or induction (0 μM FeSO4) . At 0 h, the different concentration gradients FeSO4 were added.
In conclusion, the expression of toxic gene mazF is related to the concentration of Fe2 +.And when there is enough Fe2+ in the culture environment, the expression is repressed. On the contray, when the amount of Fe2 + is not enough, the gene expression is induced so as to express toxic protein MazF to kill the bacteria.
Methods
Plasmids Construction
The promoter PfhuA1 (RBS and Fur box on it) and mazF are ligated between the parts by overlap extension PCR, and the construted fragment is ligated to pUC57 by homologous recombination. The correct construction is confirmed by sequencing.
Inoculation and cultivation
1. Methods of molecular cloning and transformation are described above. Transform this plasmid into E. coli DH5α. Then spread it onto a LB medium plates with 60 μg/mL ampicillin and incubate overnight at 37℃ in a biochemical incubator.
2. Pick three monoclonals from the same plate as parallel repeats. With the 5 ml LB medium containing 60 μg/mL ampicillin, the seed culture media are grow at 37℃ while shaking at 200 rpm overnight.
3. Take 2 mL seed culture medium into 2 mL EP tube and centrifuge for 2 minutes at 5000 rpm. Then wash the centrifugated deposit with PBS buffer to remove ferrous ions for three times. After that resuspend the deposit in the M9 medium (60 μg/ml ampicillin) and make the final volume 5 mL.
4. Measure the OD600 value of the resuspending culture media in an automatic microplate reader (SynergyH1 hybrid multimodal reader). Then dilute the resuspending culture medium to OD600 = 0.02 and add FeSO4 solution (40 mM) to the final concentration of 0 μM, 10 μM , 20 μM, 40 μM. The blank control groups are the same volume of M9 medium containing 60 μg/mL ampicillin which are added the corresponding concentration gradient FeSO4 solution respectively.
5. Incubate the cultures for 9 h at 37℃ while shaking at 200 rpm.
6. Samples are taken from each group once an hour and measured by UV spectrophotometer. And then convert the raw data to OD600.
7. Take 50uL from the seed culture medium and add it into 5ml LB medium containing 60 μg/mL ampicillin. Incubate for 8-9 hours at 37℃ while shaking at 200 rpm.
8. Dilute the incubated bacterial fluid to 102 and 103 times respectively.
9. Take 10 uL bacterial fluid from the two diluted groups, spread it onto M9 medium containing 60 μg/mL ampicillin respectively. Then two filter papers with a drop of 10 uL of FeSO4 (40 mM) and 10 uL of double distilled water respectively are put onto each plate. Blank controls are fresh M9 medium containing ampicillin (Amp, 60 μg/mL) with the engineered bacteria. Incubate them overnight at 37℃ in a biochemical incubator.
10. All experiments above are carried out in 3 biological replications.