Difference between revisions of "Part:BBa K143082"
m |
m |
||
| Line 28: | Line 28: | ||
<span class='h3bb'><big>'''Part Characterisation'''</big></span> | <span class='h3bb'><big>'''Part Characterisation'''</big></span> | ||
| + | |||
| + | [[Image:B.subtilisRFP.PNG|400px|center]] | ||
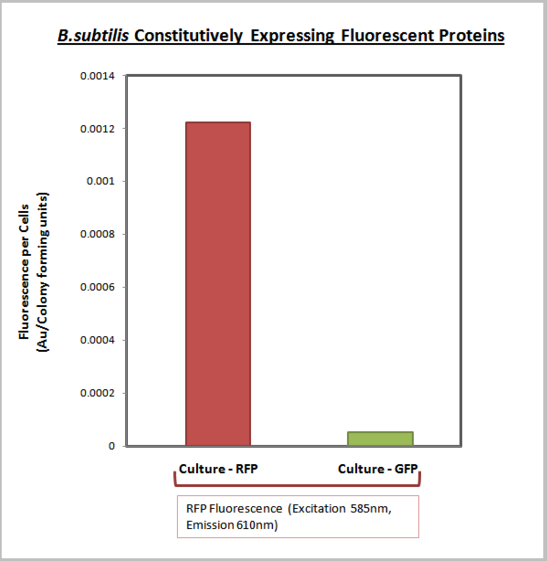
| + | This test construct was used for the characterise the expression output of the promoter pVeg combined with the ribosome binding site spoVG. The characterisation was performed using a plate reader to measure fluorescence and O.D.<sub>600</sub>. The O.D.<sub>600</sub> data was converted to cell number (colony forming units) using the calibration curve O.D.<sub>600</sub> vs cell number. Cell number was then used to normalise the fluorescence to give the final normalised units of fluorescence per cell (colony forming unit). The graph below shows the results of a ''B.subtilis'' culture containing this test construct and a culture of ''B.subtilis'' containing the test construct [https://parts.igem.org/wiki/index.php?title=Part:BBa_K143079 BBa_K143078] as a negative control. It is clear that ''B.subtilis'' containing this test construct is expressing mRFP. To learn more about the characterisation please [http://2008.igem.org/Team:Imperial_College/Biobricks click this link.] | ||
Revision as of 22:08, 29 October 2008
Pveg-spoVG RFP expression construct
RFP gene under expresson of the Pveg promoter and spoVG RBS of B. subtilis, with a chloramphenicol resistance marker for ease of selection.
Also contains the 5' and 3' amyE integration sequences to allow integration into B. subtilis at the amyE locus.
This device was used by the Imperial iGEM 2008 team to characterise the Pveg promoter and spoVG RBS (Combined part BBa_K143053) as part of the Biofabricator subtilis project.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2136
Illegal AgeI site found at 2248 - 1000COMPATIBLE WITH RFC[1000]
Part Characterisation
This test construct was used for the characterise the expression output of the promoter pVeg combined with the ribosome binding site spoVG. The characterisation was performed using a plate reader to measure fluorescence and O.D.600. The O.D.600 data was converted to cell number (colony forming units) using the calibration curve O.D.600 vs cell number. Cell number was then used to normalise the fluorescence to give the final normalised units of fluorescence per cell (colony forming unit). The graph below shows the results of a B.subtilis culture containing this test construct and a culture of B.subtilis containing the test construct BBa_K143078 as a negative control. It is clear that B.subtilis containing this test construct is expressing mRFP. To learn more about the characterisation please [http://2008.igem.org/Team:Imperial_College/Biobricks click this link.]