Difference between revisions of "Part:BBa K3332028"
| (7 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3332028 short</partinfo> | <partinfo>BBa_K3332028 short</partinfo> | ||
| − | Aminoalkylphosphonate N-acetyltransferase with a 6*histidine-tag, which can degradate AMPA. Use BBa_K823004 to construct a new part that can catalyze the acyl transfer reaction between aminomethylphosphoric acid and acetyl-CoA, his tag is used for protein purification. | + | Aminoalkylphosphonate N-acetyltransferase with a 6*histidine-tag, which can degradate AMPA. Use <partinfo>BBa_K823004</partinfo> to construct a new part that can catalyze the acyl transfer reaction between aminomethylphosphoric acid and acetyl-CoA, his tag is used for protein purification. |
| + | |||
===Biology=== | ===Biology=== | ||
Under natural conditions, many microorganisms can use the glyphosate oxidoreductase to oxidize glyphosate to aminomethylphosphoric acid (AMPA). | Under natural conditions, many microorganisms can use the glyphosate oxidoreductase to oxidize glyphosate to aminomethylphosphoric acid (AMPA). | ||
| − | Phn system is a gene cluster for organophosphorus transport and degradation in many microorganisms. The phnO gene encodes the acyltransferase which catalyzes the transfer of acyl to AMPA by acetyl-coa. Through this one-step acyl transfer reaction, AMPA can be converted into glyphosate analogue, which can then be degraded by C-P lyase. | + | |
| + | Phn system is a gene cluster for organophosphorus transport and degradation in many microorganisms. The ''phnO'' gene encodes the acyltransferase which catalyzes the transfer of acyl to AMPA by acetyl-coa. Through this one-step acyl transfer reaction, AMPA can be converted into glyphosate analogue, which can then be degraded by C-P lyase. | ||
===Usage=== | ===Usage=== | ||
| − | In order to easily purify PhnO, we added a his-tag (6*His) at the C-terminal. we ligased the strong | + | In order to easily purify PhnO, we added a his-tag (6*His) at the C-terminal. we ligased the strong promoter、RBS、Terminator(<partinfo>BBa_J23100</partinfo>、<partinfo>BBa_B0034</partinfo>、<partinfo>BBa_B0015</partinfo>) and the parts (phnO)on the expression vector pSB1C3 by standard assembly. Then the ligation mixture was transformed into ''E. coli'' DH5α & ''E. coli'' BL21 (DE3), and the correct recombinant one was confirmed by chloramphenicol, enzyme-cut identification and sequencing. |
| + | |||
| + | |||
===Characterization=== | ===Characterization=== | ||
| − | 1. Agarose Gel Electrophoresis | + | '''1. Agarose Gel Electrophoresis''' |
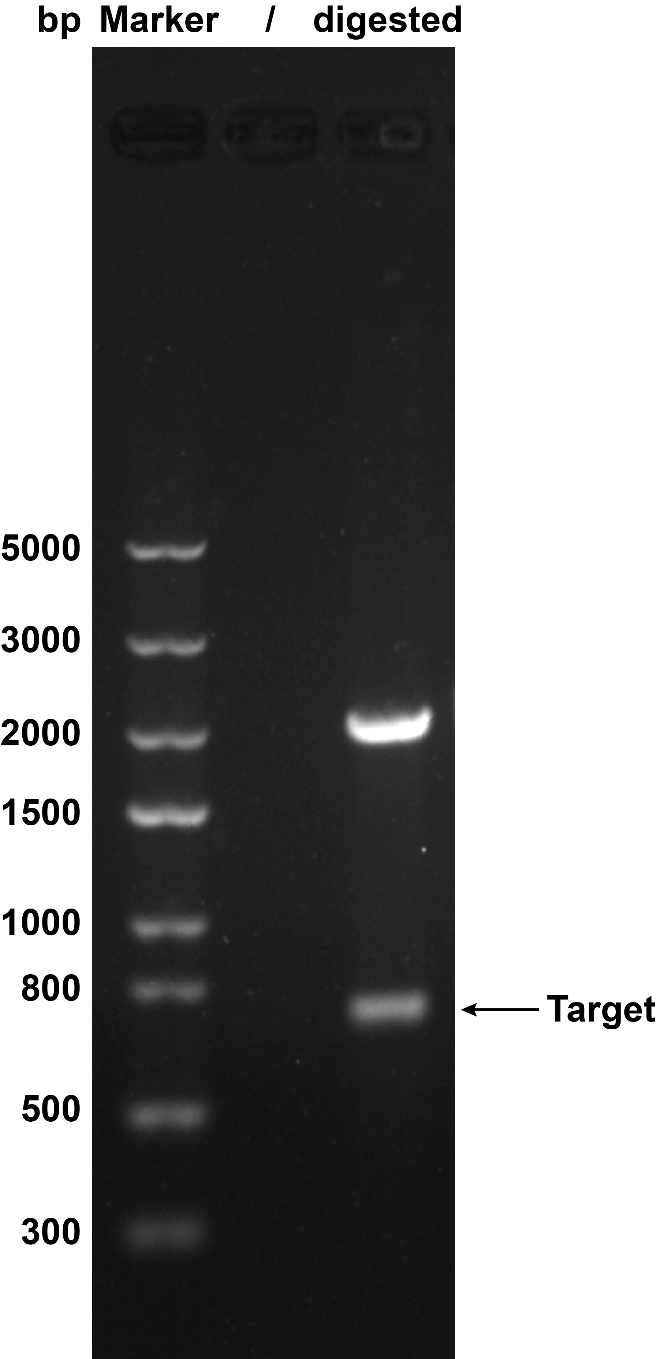
| − | When we were building this circuit, enzyme-cut identification was used to certify the plasmid was correct. We used the '' | + | |
| − | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 1.png|thumb|300px|Fig.1 The result of plasmid cut with enzyme '' | + | When we were building this circuit, enzyme-cut identification was used to certify the plasmid was correct. We used the ''Eco''R I and ''Pst'' I to cut the plasmid, then we got the target separate fragment-686bp. |
| − | 2. SDS-PAGE | + | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 1.png|thumb|300px|Fig.1 The result of plasmid cut with enzyme ''Eco''R I and ''Pst''I. Plasmid: pSB1C3.]]</th><th></table> |
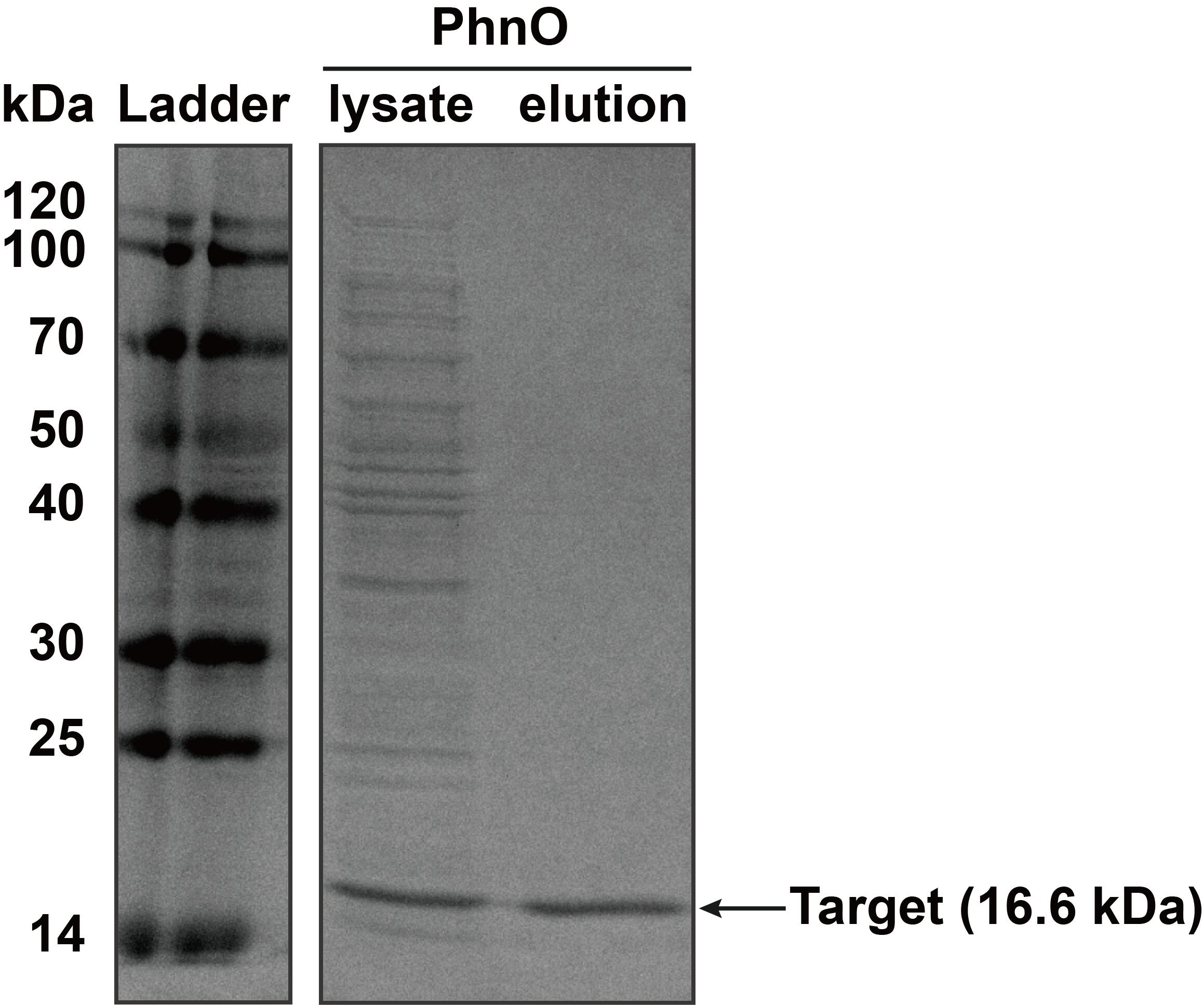
| − | The constructed plasmid was transformed into E. coli BL21 (DE3). Owing to the his-tag, the PhnO could be easily purified through AKTA with Ni-NTA. Compared with the lysate, the PhnO in the eluant can be found at around | + | |
| + | '''2. SDS-PAGE''' | ||
| + | |||
| + | The constructed plasmid was transformed into ''E. coli'' BL21 (DE3). Owing to the his-tag, the PhnO could be easily purified through AKTA with Ni-NTA. Compared with the lysate, the PhnO in the eluant can be found at around 16 kDa. Both of them were electrophoresed on a sodium dodecyl sulfate (SDS)-12% (wt/vol) polyacrylamide gel, followed by Coomassie blue staining. | ||
<table><tr><th>[[File:T--XMU-China2020--BBa K3332028 2.png|thumb|300px|Fig.2 SDS-PAGE analysis of protein in lysate of ''E. coli'' BL21 (DE3) cells and the eluant after AKTA by Coomassie blue staining. Target bands can be seen at about 16 kDa]]</th><th></table> | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 2.png|thumb|300px|Fig.2 SDS-PAGE analysis of protein in lysate of ''E. coli'' BL21 (DE3) cells and the eluant after AKTA by Coomassie blue staining. Target bands can be seen at about 16 kDa]]</th><th></table> | ||
| − | 3. Determination of enzyme kinetic constants | + | |
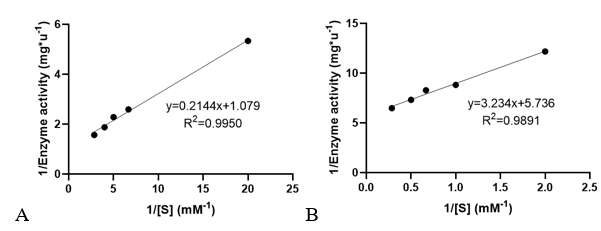
| − | Velocities for the reaction of PhnO were determined using 4,4′-dithiodipyridine (DTDP) to continuously detect the formation of the product CoA at 324 nm (thiopyridone: ε= 19800 M- | + | '''3. Determination of enzyme kinetic constants''' |
| + | |||
| + | Velocities for the reaction of PhnO were determined using 4,4′-dithiodipyridine (DTDP) to continuously detect the formation of the product CoA at 324 nm (thiopyridone: ε= 19800 M<sup>-1</sup> ·cm<sup>-1</sup> ) at 25 °C. (1) Enzyme activity was determined through enzyme-labeled instrument. | ||
| + | |||
Km and kcat of different substrates were determined with a concentration gradient. | Km and kcat of different substrates were determined with a concentration gradient. | ||
| − | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 | + | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 6.png|thumb|500px|Fig 3. The relationship of 1/Enzyme activity and 1/concentration of AcCoA and AMPA. Used to determine the enzyme kinetic constants.]]</th><th></table> |
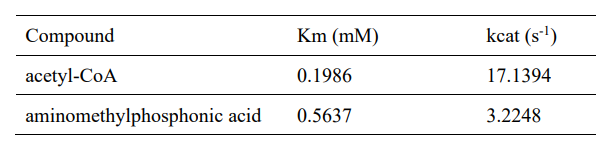
| − | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 | + | <table><tr><th>[[File:T--XMU-China2020--BBa K3332028 8.png|thumb|500px|Table 1. Enzyme kinetic constants with two substrates.]]</th><th></table><ref>1. J. C. Errey, J. S. Blanchard, Functional Annotation and Kinetic Characterization of PhnO from Salmonella enterica. Biochemistry 45, 3033-3039 (2006).</ref> |
| − | + | ===Sequence and Features=== | |
| − | + | ||
<partinfo>BBa_K3332028 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3332028 SequenceAndFeatures</partinfo> | ||
Latest revision as of 01:51, 28 October 2020
phnO-his-tag
Aminoalkylphosphonate N-acetyltransferase with a 6*histidine-tag, which can degradate AMPA. Use BBa_K823004 to construct a new part that can catalyze the acyl transfer reaction between aminomethylphosphoric acid and acetyl-CoA, his tag is used for protein purification.
Biology
Under natural conditions, many microorganisms can use the glyphosate oxidoreductase to oxidize glyphosate to aminomethylphosphoric acid (AMPA).
Phn system is a gene cluster for organophosphorus transport and degradation in many microorganisms. The phnO gene encodes the acyltransferase which catalyzes the transfer of acyl to AMPA by acetyl-coa. Through this one-step acyl transfer reaction, AMPA can be converted into glyphosate analogue, which can then be degraded by C-P lyase.
Usage
In order to easily purify PhnO, we added a his-tag (6*His) at the C-terminal. we ligased the strong promoter、RBS、Terminator(BBa_J23100、BBa_B0034、BBa_B0015) and the parts (phnO)on the expression vector pSB1C3 by standard assembly. Then the ligation mixture was transformed into E. coli DH5α & E. coli BL21 (DE3), and the correct recombinant one was confirmed by chloramphenicol, enzyme-cut identification and sequencing.
Characterization
1. Agarose Gel Electrophoresis
When we were building this circuit, enzyme-cut identification was used to certify the plasmid was correct. We used the EcoR I and Pst I to cut the plasmid, then we got the target separate fragment-686bp.
2. SDS-PAGE
The constructed plasmid was transformed into E. coli BL21 (DE3). Owing to the his-tag, the PhnO could be easily purified through AKTA with Ni-NTA. Compared with the lysate, the PhnO in the eluant can be found at around 16 kDa. Both of them were electrophoresed on a sodium dodecyl sulfate (SDS)-12% (wt/vol) polyacrylamide gel, followed by Coomassie blue staining.
3. Determination of enzyme kinetic constants
Velocities for the reaction of PhnO were determined using 4,4′-dithiodipyridine (DTDP) to continuously detect the formation of the product CoA at 324 nm (thiopyridone: ε= 19800 M-1 ·cm-1 ) at 25 °C. (1) Enzyme activity was determined through enzyme-labeled instrument.
Km and kcat of different substrates were determined with a concentration gradient.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 159
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
- ↑ 1. J. C. Errey, J. S. Blanchard, Functional Annotation and Kinetic Characterization of PhnO from Salmonella enterica. Biochemistry 45, 3033-3039 (2006).