Difference between revisions of "Part:BBa K2306010"
(→iGEM 2020 QHFZ-China, improvement (For Gold)) |
(→iGEM 2020 QHFZ-China, improvement (For Gold)) |
||
| Line 141: | Line 141: | ||
<h3><b>Design & Improvement</b></h3> | <h3><b>Design & Improvement</b></h3> | ||
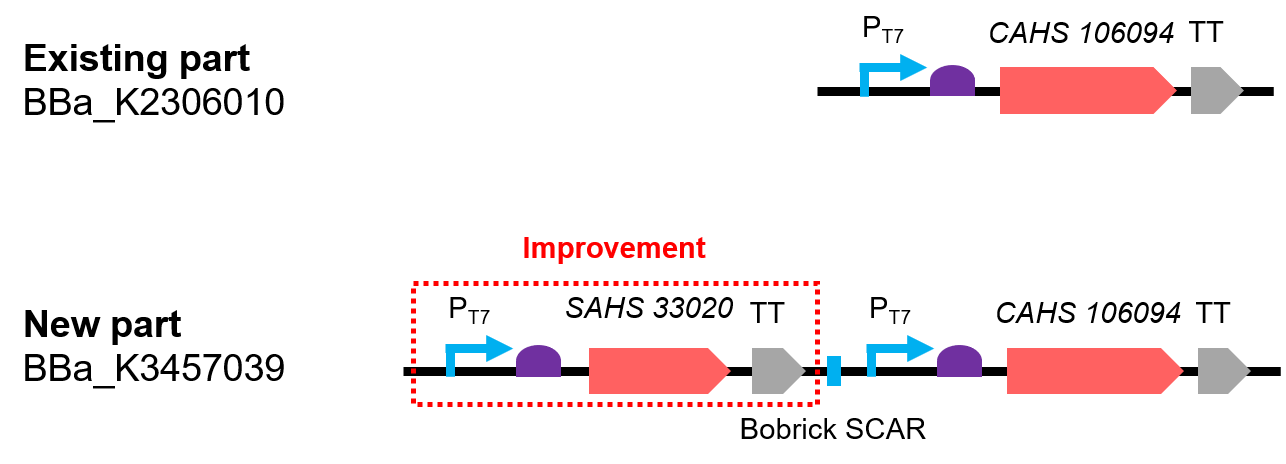
[[File:T--QHFZ-China--330-106-1.png|600px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct.]] | [[File:T--QHFZ-China--330-106-1.png|600px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct.]] | ||
| − | <p style="clear:left;"> The new part is a combination by two expression module and the two expressed | + | <p style="clear:left;"> The new part is a combination by two expression module and the two expressed proteins combined has a synergistic effect (Fig. 1). The part was constructed by biobrick ligation. We incorporated SAHS 33020 module into the vector first, then cut the vector by <i>SpeI and </i>PstI, cut the CAHS 106094 module by <i>XbaI</i> and <i>PstI</i>, at last used T4 ligase to ligate the two fragments. </p> |
| − | <p>Compared with the existing art, | + | <p>Compared with the existing art, this new part can express one more useful protein. In addition, we also added Olac sequence after the T7 promoter, so that the expression could be controlled by adding iPTG; we added a 6x His tag sequence at the 5' end of the CDS of the two protein for convenient detection by Western Blot.</p> |
<h3><b>Documentation:</b></h3> | <h3><b>Documentation:</b></h3> | ||
<h4><b>Introduction: </b></h4> | <h4><b>Introduction: </b></h4> | ||
| − | <p style="clear:left;"> This year, we tried to introduce a new | + | <p style="clear:left;"> This year, we tried to introduce a new bio-preservation method. We used freeze-drying to make the engineered bacteria into dry powder. Then the powder can be stored at room temperature for a long time. This method can make refrigeration obsolete in bacteria storage, which will promote the practical application of engineered bacteria outside of the laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freezing, drying and being in a vacuum, are lethal to bacteria. We use TDPs to help bacteria survive these extreme conditions. In the past, when studying cells, scientists attempted drying live cells, so we are the first to study freeze-drying live cells.</p> |
| − | <p style="clear:left;"> & | + | <p style="clear:left;">  However, though a single unit of TDP could enhance the survival rate of bacteria during the freeze-drying process and subsequent dry storage, the survival rate should be elongated. As far as we knew from Human Practices, the current survival rate of bacteria after freeze-drying is around 10%, even great protective agents are added and advanced freeze-drying protocol is used. We can imagine that on this basis, if single TDP is added, the survival rate would be lengthened. However, it is not very realistic to reach more than 50%. As a result, the method (adding TDP) should be further improved.</p> |
[[File:T--QHFZ-China--330-106-3.png|600px|thumb|left|Figure 2. Co-expressing SAHS 33020 and CAHS 106094 to protect engineered bacteria.]] | [[File:T--QHFZ-China--330-106-3.png|600px|thumb|left|Figure 2. Co-expressing SAHS 33020 and CAHS 106094 to protect engineered bacteria.]] | ||
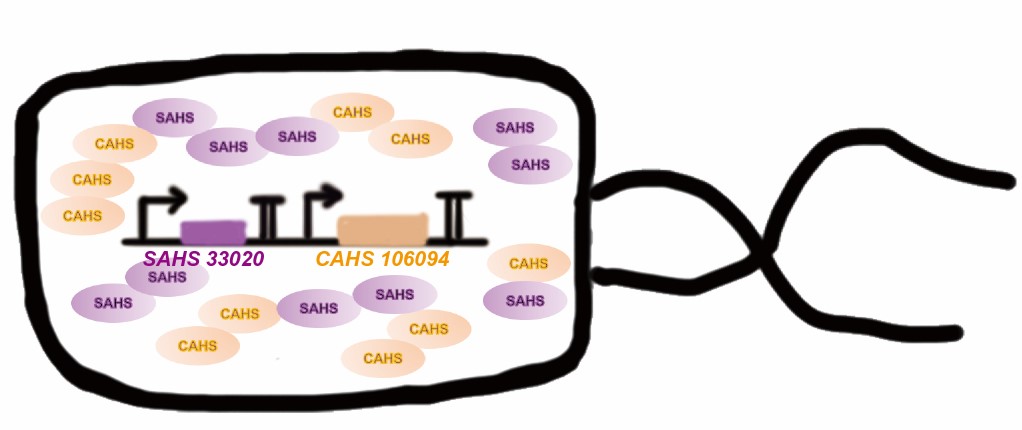
<p style="clear:left;"> Tardigrade co-expresses many TDPs to survive the drought. The proteins may have some supplementary effect, because the structures are not the same. However, no one studied co-expressing them before. Here we tried to combine two TDPs (Fig. 2) and get a better result than expressing single TDP.</p> | <p style="clear:left;"> Tardigrade co-expresses many TDPs to survive the drought. The proteins may have some supplementary effect, because the structures are not the same. However, no one studied co-expressing them before. Here we tried to combine two TDPs (Fig. 2) and get a better result than expressing single TDP.</p> | ||
| Line 162: | Line 162: | ||
(2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br> | (2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br> | ||
(3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.<br> | (3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.<br> | ||
| − | (4) | + | (4) Re-suspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.<br> |
(5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.<br> | (5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.<br> | ||
(6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.<br> | (6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.<br> | ||
| Line 174: | Line 174: | ||
【Day 5】Cell Count<br> | 【Day 5】Cell Count<br> | ||
(1) Take out the LB plate and take photos to record experimental results.<br> | (1) Take out the LB plate and take photos to record experimental results.<br> | ||
| − | (2) Use the automatic cell counting function of Image J to count the | + | (2) Use the automatic cell counting function of Image J to count the clone number on the LB plate, then compare the results between each group. |
</p> | </p> | ||
<h4><b>Results:</b></h4> | <h4><b>Results:</b></h4> | ||
Latest revision as of 17:58, 27 October 2020
Cytosolic-abundant heat soluble protein 106094 "CAHS 106094" (T7 promoter, RBS and double terminator
This composite BioBrick features the basic part BBa_k2306005 that codes for the production of the tardigrade intrinsically disordered protein (TDP) "CAHS 106094". This is one of the several heat soluble proteins found in the tardigrades capable of protecting other fragile biological parts upon desiccation by generating a crystal matrix. Therefore, this tardigrade protein can be combined with other perishable biological material (e.g. proteins) to increase its stability after dehydration or even enable the possibility of stabilizing material with just a previous drying step. The sequence encoding for the gene is preceded by a T7 promoter BBa_I712074 and an RBS BBa_K2306014, which allows inducible overexpression of the protein with IPTG; and followed by a double terminator consisting of a rrnB T1 terminator and T7Te terminator BBa_B0015. This was the part employed in TU Delft's project for the expression and production of CAHS 106094. More information about our project can be found on our [http://2017.igem.org/Team:TUDelft/Results#TDP results page].
Navigation
iGEM_17 TU DelftiGEM_20 QHFZ-China
Tardigrade-specific Intrinsically Disordered Proteins (TDPs) by TU Delft 2017
Tardigrade-specific intrinsically disordered proteins (TDPs) are a set of intrinsically disordered proteins found in tardigrades, a series of microscopic animals known for their ability to survive in the harshest conditions, to the extent of even being able to survive in the space (Simon et al. 2014). Recently, Boothby et al. (2017) discovered that the unmatched resilience of the tardigrades towards desiccation was caused by two families of TDPs: the cytosolic and secretory abundant heat-soluble proteins (CAHS and SAHS respectively), named after their occurrence in the tardigrades; being CAHS proteins most commonly found in the cytosol and SAHS proteins found in the culture medium during the desiccation process (Yamaguchi et al. 2012).These intrinsically disordered proteins (i.e. proteins lacking a tertiary structure) are produced by the tardigrades upon hydric stress or desiccation and generate a glass-like matrix that substitutes water and protects cellular components. This matrix redissolves upon rehydration, thus leaving behind undamaged functional cells(Sloan et al. 2017). For the study of this TDPs, we chose the proteins with the highest impacts on the survival rates, namely CAHS 94205, CAHS 106094, SAHS 68234 and SAHS 33020 according to the results by Boothby et al. (2017).
Identification of the TDPs
After isolation of the four TDPs according to the protocol described by Boothby et. al. (2017), the proteins were identified by SDS-PAGE and mass spectrometry and their resulting concentration was determined to range from 1 to 3 g/L by Bradford Assay (using bovine serum albumin as standards).
SDS-PAGE

Figure 1:SDS-PAGE gels of TDPs after protein purification. Gel a features the ladder (L), CAHS 94205 (#1), SAHS33025 (#2) and the control with the backbone PSB1C3 (Blank) while gel b includes CAHS 106094 (#3), the ladder (L) and SAHS 68234 (#4)
We carried out an SDS-PAGE for all four proteins following the [SDS-PAGE protocol]. Additionally, we included a protein solution resulting from the purification of the bacterial strain featuring the same backbone as our BioBricks. As depicted in Figure 1, the samples from the purified TDPs led to a series of additional bands absent in the control, indicating that we successfully produced and isolated all four tardigrade-specific proteins.
Mass spectrometry
In addition to SDS-PAGE of the purified protein solutions, the identity of the proteins was also confirmed by mass spectrometry. To do this, a sequence unique to each TDPs’ amino acid sequence were chosen and screened for their presence in the proteome of the expression strain, BL21 (DE3). Subsequently, four cell cultures expressing one TDP each were used to screen for all chosen sequences by MS and an additional culture only featuring the same backbone (PSB1C3) as a control.

Figure 2: Bar chart graph including the peak areas of the TDP samples analyzed by mass spectrometry after purification. For the control, the expression strain only contained the same backbone.
The peak areas of the resulting mass spectrographs shown in Figure 2 reflect the occurrence of a given sequence in the sample. The unique peptides that were screened for were only present in each TDP expected to contain the sequence. The differences in peak height in Figure 2 can be attributed to the different lengths of the target peptides which influence the readout due to the variation of the mass/charge ratios. Hence, it can be concluded that the results were positive and the identity of the proteins could be verified by mass spectrometry.
Bacteria desiccation protection
As described by Boothby et al. (2017), the production of tardigrade-specific proteins by heterologous organisms increases their desiccation tolerance significantly. Therefore, to the end of further characterizing our BioBricks, iGEM Wageningen was asked to carry out experiments with our plasmids to test the increase in bacteria desiccation tolerance by CAHS94205, CAHS106094 and SAHS33020, as part of their collaboration with TU Delft iGEM 2017. However, for the sake of gaining a better understanding of the proteins’ function, the Wageningen iGEM team performed additional experiments by washing with PBS buffer, a procedure that had not been done before.

Figure 3: LDH activity after drying and subsequent rehydration for the different TDPs.
As seen in Figure 3, the number of colonies found after drying and rehydrating the bacteria (BL21(DE3)) is considerably larger than that for the empty vector of the same bacterial strain (1/103-104). This proves that the survival of bacteria is greatly enhanced by the presence of the three TDPs in the cells which complies with the results from Boothby et al. (2017). However, a closer look at the differences in trend between the samples washed with water and PBS reveals an increase in the survival of bacteria with plasmids for the production of cytosolic abundant proteins (CAHS) and a decrease in the secretory abundant protein SAHS 33020. It is possible that since a PBS solution is more similar to the cytosol, which is where CAHS proteins are mostly found, it is better suited to the CAHS. Likewise, as SAHS proteins have been mostly found in the culture medium, the colony decrease for the vector including SAHS might not occur in media of that composition.
Lactate Dehydrogenase enzyme protection
To the end of assessing the protective effect of TDPs on non-native proteins, all tardigrade-specific proteins were tested as buffer excipients (in concentrations ranging from 0.1 to 1 g/L) for the enzyme Lactate Dehydrogenase (LDH) in an enzymatic activity assay based on that described by Boothby et al. (2017) and Goya et al. (2015).All measurements were performed in triplicates.

Figure 4: LDH activity after freeze-drying and drying with TDPs. LDH activity after freeze-drying (FD)/ drying with different concentrations for (a) CAHS 94205 and (b) SAHS 33020 and rehydrating as a function of time. Experiments realized at room temperature.
As depicted in Figure 4, the LDH activity is least preserved at the lowest TDP concentration for all proteins and a maximum of activity preserved for the highest concentrations assessed, with the exception of CAHS 94205, though the error bars should be considered. This indicates that the higher the protein concentration in the solution, the better the activity preserved, and thus the better the protection of the enzyme. Such a trend was also observed in our [lattice model] and by Boothby et. al. (2017) thus substantiating the validity of our results.
Furthermore, the preserved most enzymatic activity at the highest concentration studied (1 mg/mL), did not differ between the four proteins. It should still be noted that, unlike the other proteins, CAHS 106094 did not show a remarkable preservation of activity at a concentration of 0.1 mg/mL and a much lower preservation at a concentration of 0.5 g/L in comparison to the other TDPs.
In view of the previous results, we adapted the LDH assay previously carried out with CAHS 94205 and SAHS 33020 to test the protective capability of the proteins for 18 days and thus evaluate the use of the TDPs for long-term storage at room temperature. Samples similar to those of the previous procedure were prepared and stored dried at room temperature for different amounts of time before their resuspension and measurement.

Figure 5: Bacteria desiccation tolerance with TDPs. Amount of colonies remaining after desiccation and rehydration for the empty vector, TU Delft’s BioBricks for the production of CAHS 94205 , CAHS 106094 and SAHS 33020 with BL21(DE3). Experiments performed by iGEM Wageningen UR.
As shown in Figure 5, LDH maintains its activity when stored dried with the cytosolic and secretory TDPs, while its activity was not preserved when freeze-dried or dried in the absence of TDPs. Furthermore, it can be seen in Figure 5 that for TDP concentrations of 0.1 mg/mL only SAHS 33020 could maintain a significant amount of activity after a day. Although both proteins successfully maintained LDH activity at higher TDP concentrations, the activity decrease over time was substantially lower at a concentration of 1 mg/mL compared to that observed for 0.5 mg/mL. Moreover, for a concentration of 1 mg/mL, it can be concluded that the variations in activity over time for SAHS 33020 are lower than those for CAHS 94205, suggesting that the mechanism by which the SAHS proteins confer their protection is more effective for long periods of time. Our assays reveal that LDH in combination with SAHS 33020 is still active after being stored dry for 18 days at room temperature. Nevertheless, the activity drops in the data discussed above could also be due to other factors than just the limitations of the proteins. It should be noted that after drying the samples no additional measures to ensure the dryness of the samples were taken as these were kept in a drawer instead of in a desiccator. Moreover, interactions with any air components should not be neglected either, as the samples were not purged with an inert gas prior to their storage.
Cas13a protection
After evaluating the influence of TDPs on the desiccation tolerance of LDH, an established procedure, we wanted to assess their protective performance with Cas13a. Our [lattice model] indicated that it was best to use CAHS 94205 and SAHS 33020 to preserve Cas13a activity at a concentration of 1 g/L or higher. Both CAHS 94205 and SAHS 33020 were dried with Cas13a in a concentration of 1 g/L, as the protein purification yielded low concentrations for SAHS 33020. Subsequently, Cas13a was rehydrated and its activity measured via an [RNase alert fluorescence assay]. All measurements were performed in duplicate.
As shown in Figure 6, the fluorescence intensity triggered by the RNase-like activity of Cas13a dried without any TDPs is very low in comparison to that of the Cas13a undried and stored frozen. However, while the fluorescence intensity and thus Cas13a activity was higher when Cas13a was dried with CAHS 94205, it also seemed to lose its specificity: the fluorescence intensity after one hour was the same with crRNA and target as without.

Figure 6: Cas13a activity with CAHS 94205 in RNase Alert assay. Fluorescence intensities over time triggered by RNase activity of Cas13a both before and after drying and in combination with the crRNA and target RNA and the TDP CAHS 94205.
In view of such unexpected results and the fact that iGEM LMU & TU Munich was also working with Cas13a and similar fluorescence assays, we sent them a newly purified batch of CAHS 94205, to determine whether the same phenomenon would be observed in experiments under the same conditions.

Figure 7: Cas13a activity with CAHS 94205 in RNase Alert Assay by Munich. Fluorescence intensities over time triggered by RNase activity of Cas13a after drying with the TDP CAHS 94205, with and without crRNA and target RNA. Data from iGEM LMU & TU Munich 2017.
The results of iGEM LMU & TU Munich 2017 also showed the unexpected RNase activity of Cas13a. We, therefore, hypothesize that the conformational change in Cas13a, normally caused by the binding of the crRNA with the target, is induced as a result of an interaction between CAHS 94205 and Cas13a upon drying and resuspension. Consequently, CAHS 94205 was ruled out as a potential desiccation tolerance mediator for Cas13a.
However, when the same experiments were repeated with SAHS 33020 instead, Cas13a remained active after being dried and rehydrated, and the fluorescence intensity observed in the presence of the target and crRNA was clearly higher than in its absence (see Figure 8). This indicates that SAHS 33020 can preserve Cas13a activity after desiccation and rehydration, thus keeping the protein functional and opening the possibility of the storage of Cas13a with TDPs.

Figure 8: Cas13a activity with SAHS 33020 in RNase Alert Assay. Fluorescence intensities over time triggered by RNase activity of Cas13a after drying with the TDP SAHS 33020, with and without crRNA and target RNA.
Conclusions
We analysed the desiccation tolerance conferred to our Cas13a by our purified TDPs CAHS 94205 and SAHS 33020 and discovered that while both CAHS 94205 and SAHS 33020 preserved Cas13a’s RNase-like activity after drying, CAHS 94205 could not preserve its specificity, as the dried Cas13a in combination with the latter showed RNase activity even in the absence of the target RNA. This previous extraordinary find was furthermore corroborated by iGEM LMU & TU Munich 2017 when the same trend was observed under similar experimental conditions with a Cas13a protein originating from a different strain. Nonetheless, the results were positive for the tardigrade-specific protein SAHS 33020, as both the activity and specificity of Cas13a could be considerably preserved after drying and rehydrating. It should not be disregarded that the best preservation of activity in the [long-term assays] was achieved for SAHS 33020 as well. In future experiments, the long-term evolution of the Cas13a activity should be assessed for SAHS 33020 and other tardigrade-specific SAHS proteins as, in view of the long-term results for the TDPs with LDH, these proteins hold great potential in simplifying the storage, usage, and shipment of the many fragile chemicals and biological materials.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
iGEM 2020 QHFZ-China, improvement (For Gold)
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
This biological is the expression sequence of Cytosolic-abundant heat soluble protein 106094, also called CAHS 106094. It is a kind of Tardigrade intrinsically Disordered Protein (TDP). It is a heat soluble protein found from Tardigrade Hypsibius dujardini in 2017 [1]. Tardigrade, also called water bear, is a kind of tenacious organism. It can survive extreme environment, such as desiccation, freeze and vacuum. The super capacity of Tardigrade partially owes to TDPs. Here we found that expressing this protein can help bacteria survive the freeze-drying process and then the resultant dry bacteria powder can be stored for a long time at room temperature.
This year, we measured this part,so we would add some new data here. At the same time, we improved the part to achieve a better function. The new part is BBa_K3457039. We put the data of both parts below.
Design & Improvement
The new part is a combination by two expression module and the two expressed proteins combined has a synergistic effect (Fig. 1). The part was constructed by biobrick ligation. We incorporated SAHS 33020 module into the vector first, then cut the vector by SpeI and PstI, cut the CAHS 106094 module by XbaI and PstI, at last used T4 ligase to ligate the two fragments.
Compared with the existing art, this new part can express one more useful protein. In addition, we also added Olac sequence after the T7 promoter, so that the expression could be controlled by adding iPTG; we added a 6x His tag sequence at the 5' end of the CDS of the two protein for convenient detection by Western Blot.
Documentation:
Introduction:
This year, we tried to introduce a new bio-preservation method. We used freeze-drying to make the engineered bacteria into dry powder. Then the powder can be stored at room temperature for a long time. This method can make refrigeration obsolete in bacteria storage, which will promote the practical application of engineered bacteria outside of the laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freezing, drying and being in a vacuum, are lethal to bacteria. We use TDPs to help bacteria survive these extreme conditions. In the past, when studying cells, scientists attempted drying live cells, so we are the first to study freeze-drying live cells.
 However, though a single unit of TDP could enhance the survival rate of bacteria during the freeze-drying process and subsequent dry storage, the survival rate should be elongated. As far as we knew from Human Practices, the current survival rate of bacteria after freeze-drying is around 10%, even great protective agents are added and advanced freeze-drying protocol is used. We can imagine that on this basis, if single TDP is added, the survival rate would be lengthened. However, it is not very realistic to reach more than 50%. As a result, the method (adding TDP) should be further improved.
Tardigrade co-expresses many TDPs to survive the drought. The proteins may have some supplementary effect, because the structures are not the same. However, no one studied co-expressing them before. Here we tried to combine two TDPs (Fig. 2) and get a better result than expressing single TDP.
Protocol:
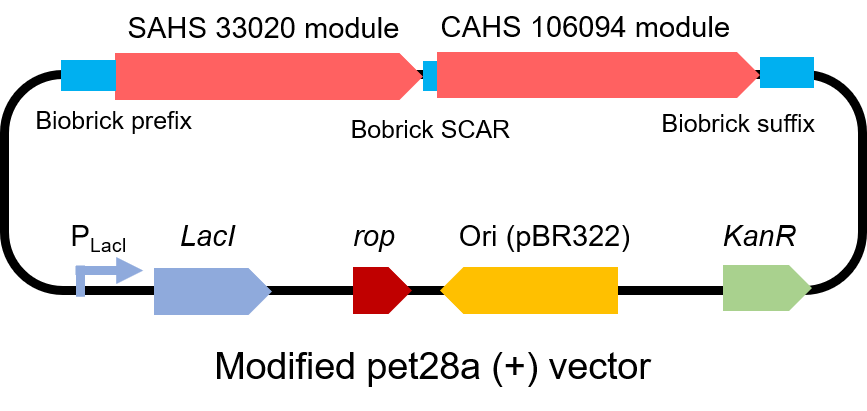
To test the effect of this part, we modified a frequently and widely used vector, pet28a+ and put this part into it (Fig. 3). Then we transformed the plasmid into E. coli BL21 strain.
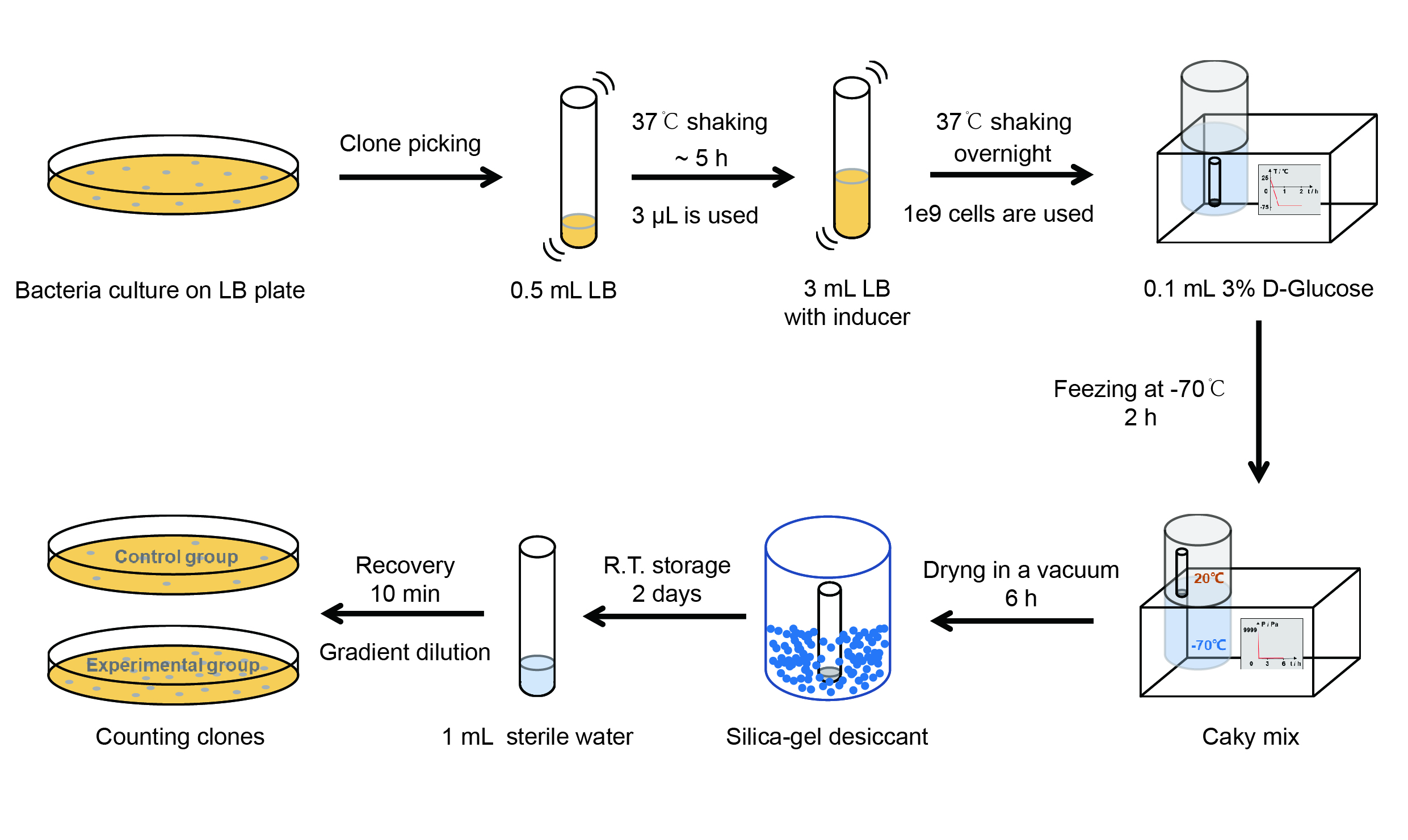
Then we used the following protocols to verify its function (Fig. 4):
【Day 1】Induction culture
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2 mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
【Day 2】Freeze-dried
(1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the experiment.
(2) Use spectrophotometer to measure the OD600 of the bacteria solution, OD600 = 1 equals to 109 cells. If the OD600 value is between 0.1 and 1, There is a linear relationship between OD600 and bacterial density. Calculate the volume of bacterial solution for 109 cells by using the formula V = 100 / (OD600 × Dilution ratio).
(3) Take out a measured amount of 109 cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.
(4) Re-suspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.
(5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.
(6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.
(7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room temperature.
【Day 3】Room temperature storage
【Day 4】Detect the survival rate
(1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.
(2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on the LB plate.
(3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several gradient dilutions.
(4) Culture the bacteria overnight at 37℃.
【Day 5】Cell Count
(1) Take out the LB plate and take photos to record experimental results.
(2) Use the automatic cell counting function of Image J to count the clone number on the LB plate, then compare the results between each group.
Results:
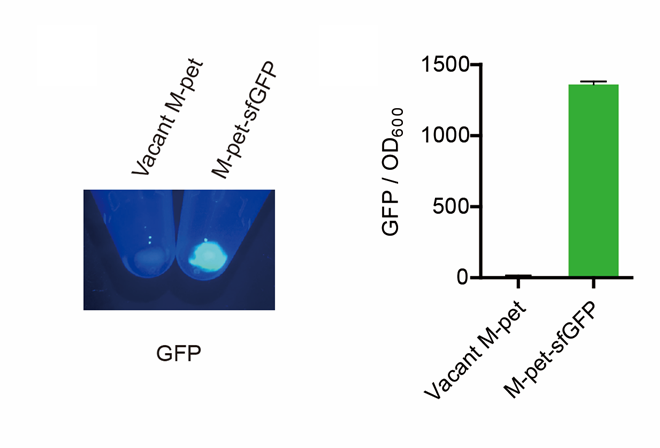
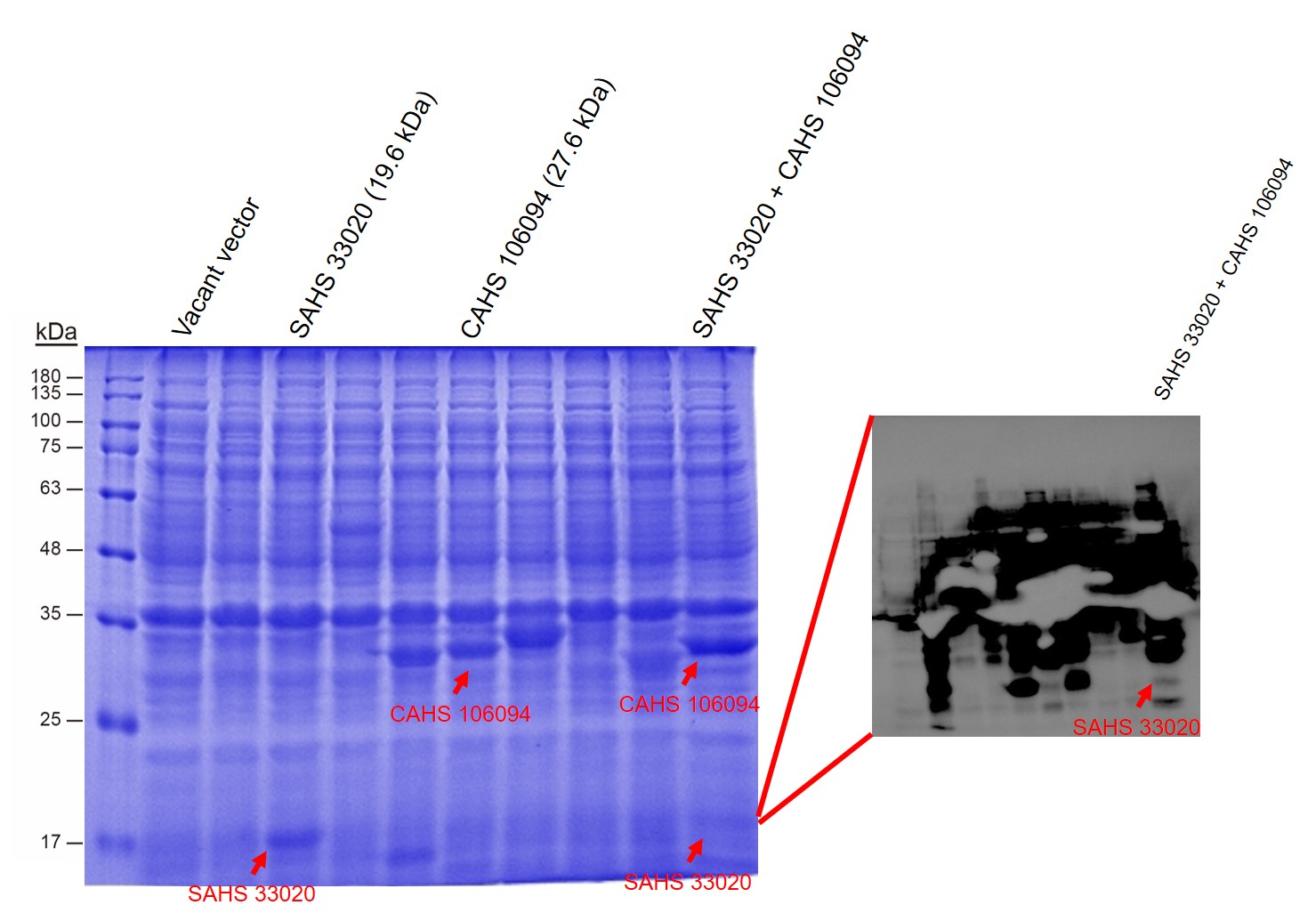
First, by a reporter, sfGFP, we confirmed that the plasmid can normally expressed exogenous proteins in E. coli BL21 strain (Fig. 5). Via SDS-PAGE, we observed the band of CAHS 106094, further verified the successfully expression (Fig.6).
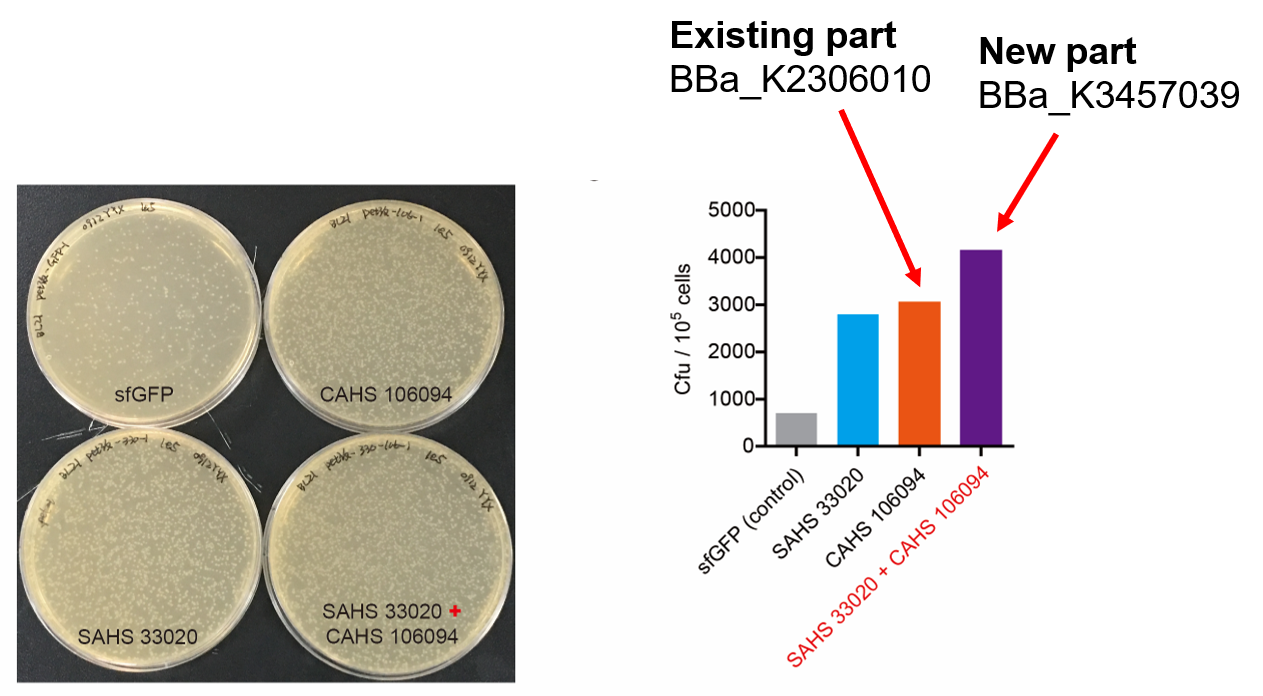
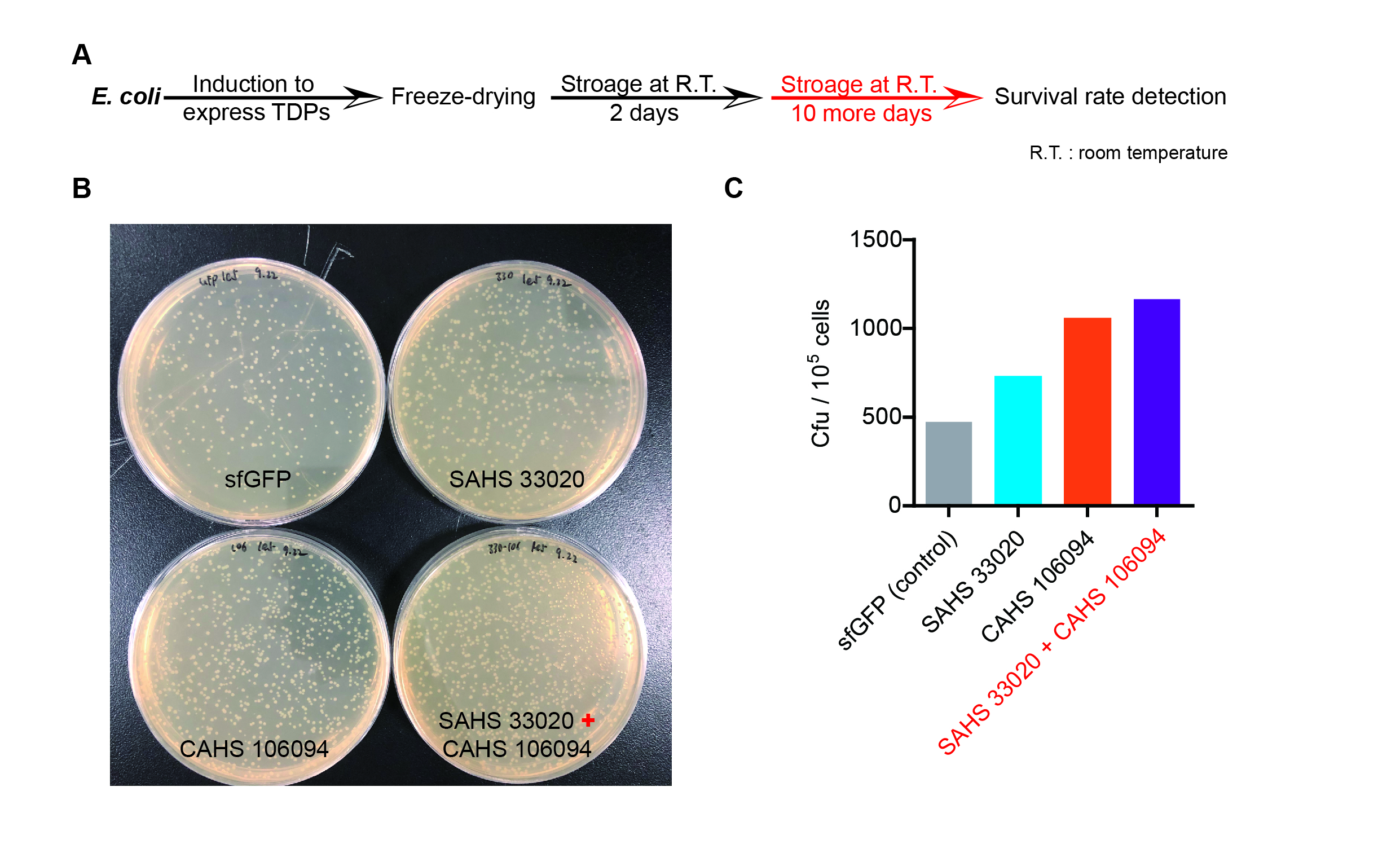
Then, by freeze-drying and recover experiment, we found that compared with the control group (sfGFP), the bacteria expressing SAHS 33020 or CAHS 106094 had a better survival rate, while co-expressing the two proteins gave a better survival rate than that of expressing single TDP (Fig. 6).
The dry storage time is 2 days, which is a litte short. So we waited for another 10 days to test if TDPs work in long-term storage. In accord with the result above, co-expressing two TDPs gave the best survival rate. (Fig. 6).
Summary:
CAHS 106094 helps the bacteria survive freeze-drying and subsequent dry storage. Additional SAHS 33020 can collaborate with CAHS 106094 and gave a bettereffect. To achieve that, you need only to transform the sequence into your bacteria.
Reference:
[1] Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., Rebecchi, L., Pielak, G.J., Koshland, D., and Goldstein, B. (2017). Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 65, 975-984 e975.