Part:BBa_K3457039
T7-RBS-SAHS 33020-T7-RBS-CAHS 106094
This biological part is the expression sequence of the cytosolic-abundant heat soluble protein 106094 (CAHS 106094) and secreted abundant heat soluble protein 33020 (SAHS 33020). They are two types of tardigrade intrinsically Disordered Proteins (TDPs), which are heat soluble proteins found from Tardigrade Hypsibius dujardini in 2017 [1]. Tardigrades, also called water bears, are incredibly tenacious organisms. They can withstand extreme environments such as desiccation, freezing, and even vacuum space. The extreme resilience of tardigrades is partly because of the TDPs. We found that expressing CAHS 106094 or SAHS 33020 can help bacteria survive the freeze-drying process, allowing the dry bacteria powder to be stored for a long time at room temperature. Moreover, co-expression of the two proteins using this part also gives a better protective effect.
Contribution
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
Design & Improvement
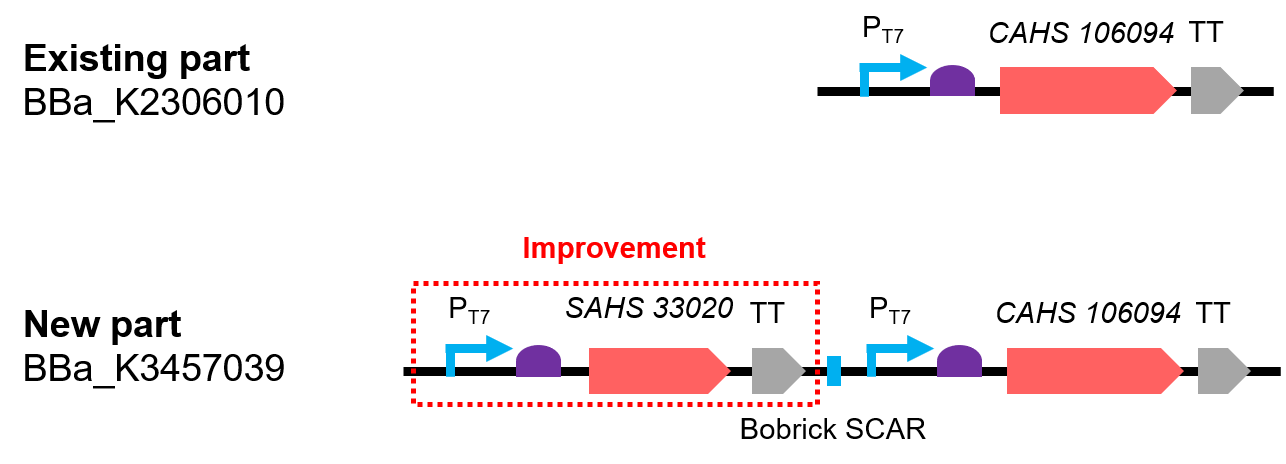
This part is an improvement from the existing part BBa_K2306010.
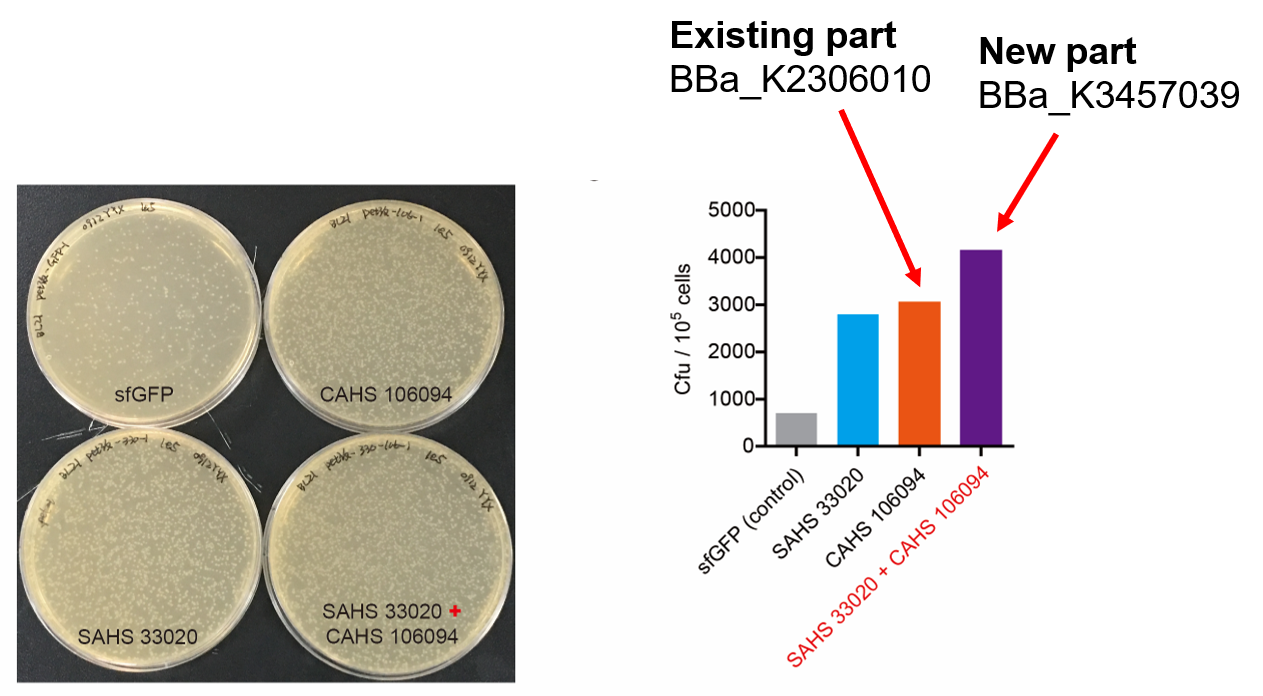
This part combines two expression modules, which gave rise to a synergistic effect (Fig. 1). The part was constructed by biobrick ligation. We constructed SAHS 33020 module into the vector first, then cut the vector by SpeI and PstI, cut the CAHS 106094 module by XbaI and PstI. To end, we used T4 ligase to ligate the two fragments.
Compared to the existing parts, it can express one more useful protein. In addition, we also added a lacO sequence after the T7 promoter so that the expression could be controlled by adding iPTG; we added a 6x His tag sequence at the 5' end of the CDS of the two proteins for convenient detection by western Blot.
Documentation:
Introduction:
This year, we tried to introduce a new bio-preservation method. We used freeze-drying to turn the engineered bacteria into a dry powder, which allows the powder to be stored at room temperature for a long time. This method renders refrigeration obsolete in bacteria storage, which facilitates practical applications outside of the laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freeze, dry, and vacuum, are lethal to bacteria. We used TDPs to help bacteria survive these extreme conditions. In the past, for studies with live cells, almost all researchers studied drying live cells, so we may be the first to study freeze-drying live cells.
However, though single TDPs could enhance the survival rate during the freeze-drying process and subsequent dry storage, the survival rate should be further enhanced. As far as we know from Human Practices, the current survival rate of bacteria after freeze-drying is around 10%, even after using protective agents and advanced freeze-drying protocols. We can imagine that if a single TDP is added, the survival rate would be enhanced. However, it is not very realistic to reach 50% or higher. As a result, the method (adding TDP) should be further improved.
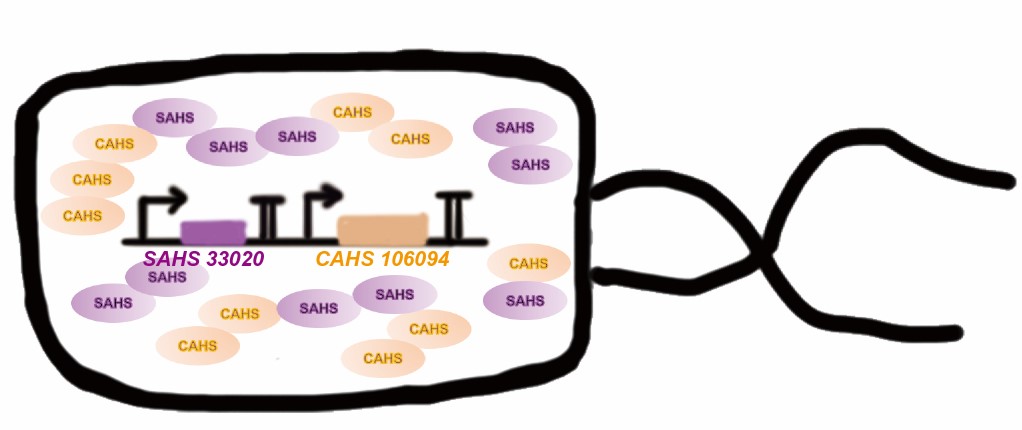
Tardigrade co-expresses many TDPs to survive the drought. The proteins may have some supplementary effect, because the structures are not the same. However, no one studied co-expressing them before. Here we tried to combine two TDPs (Fig. 2) and get a better result than expressing single TDP.
Protocol:
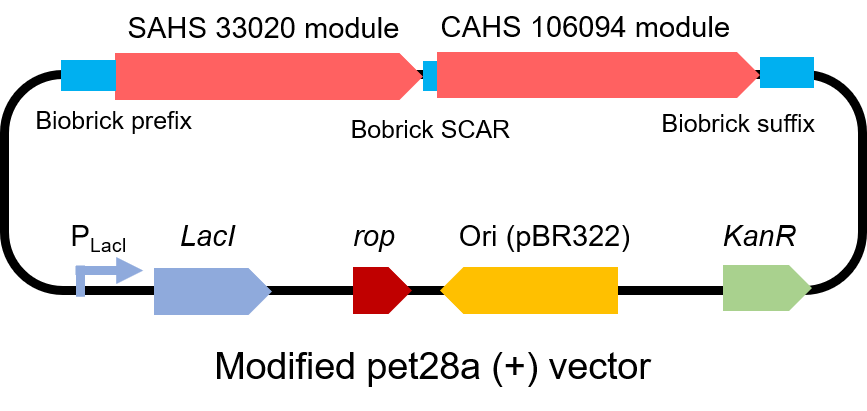
To test the affect of this part, we modified a frequently and widely used vector, pet28a+ and put this part into it (Fig. 3). Then we transformed the plasmid into E. coli BL21 strain.
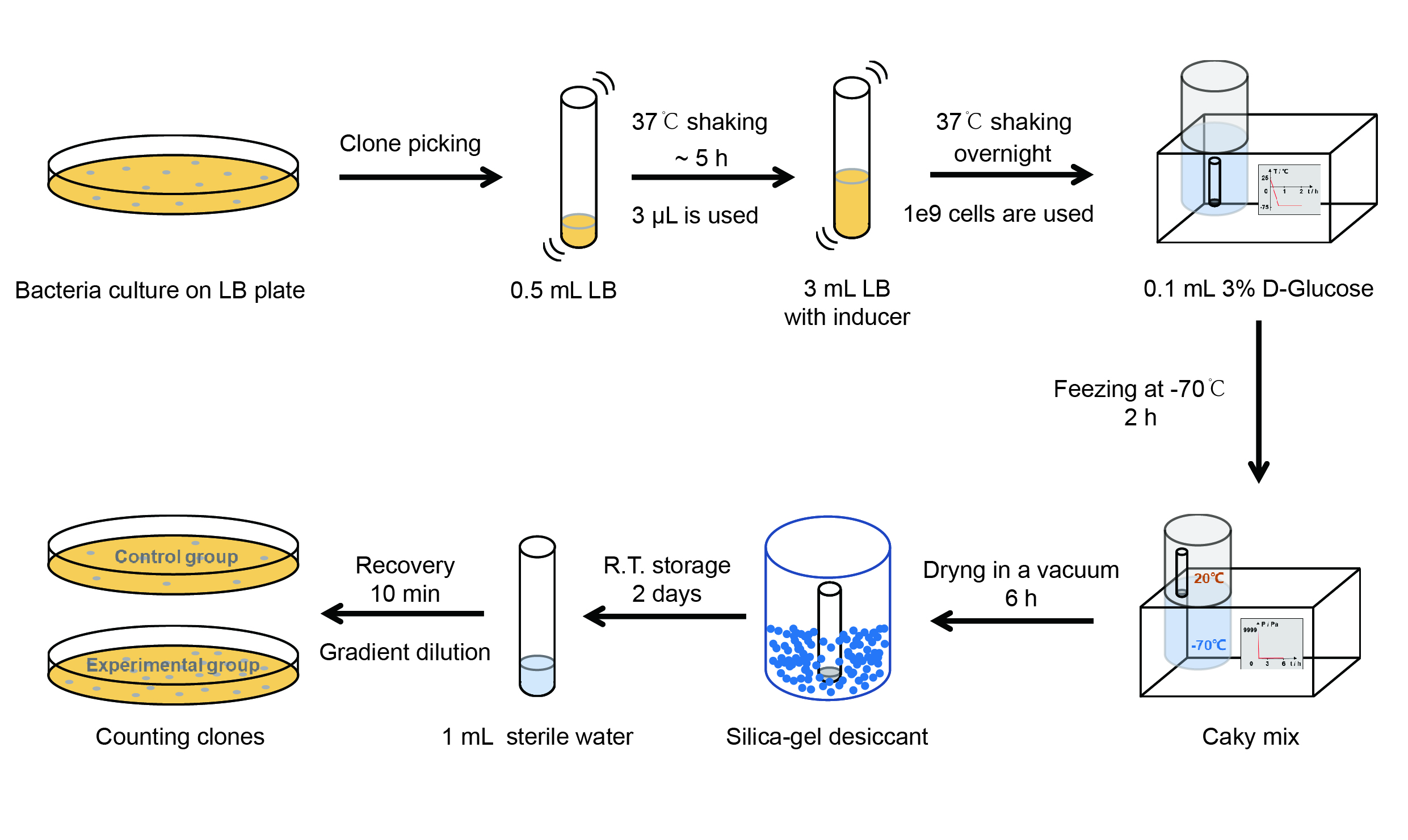
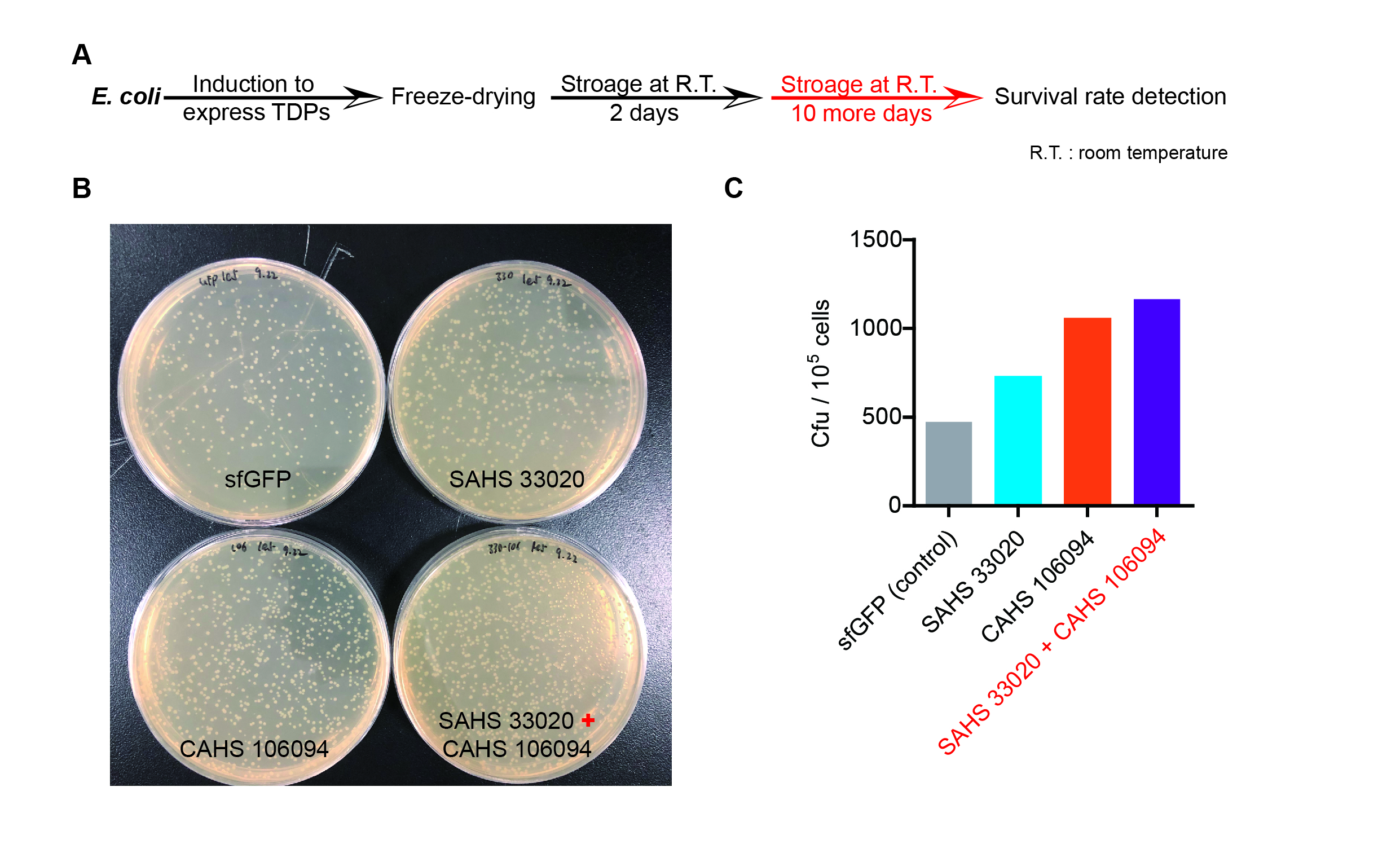
Then we used the following protocols to verify its function (Fig. 4):
【Day 1】Induction culture
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2 mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
【Day 2】Freeze-dried
(1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the experiment.
(2) Use spectrophotometer to measure the OD600 of the bacteria solution, OD600 = 1 equals to 109 cells. If the OD600 value is between 0.1 and 1, There is a linear relationship between OD600 and bacterial density. Calculate the volume of bacterial solution for 109 cells by using the formula V = 100 / (OD600 × Dilution ratio).
(3) Take out a measured amount of 109 cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.
(4) Re-suspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.
(5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.
(6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.
(7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room temperature.
【Day 3】Room temperature storage
【Day 4】Detect the survival rate
(1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.
(2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on the LB plate.
(3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several gradient dilutions.
(4) Culture the bacteria overnight at 37℃.
【Day 5】Cell Count
(1) Take out the LB plate and take photos to record experimental results.
(2) Use the automatic cell counting function of Image J to count the clone number on the LB plate, then compare the results between each group.
Results:
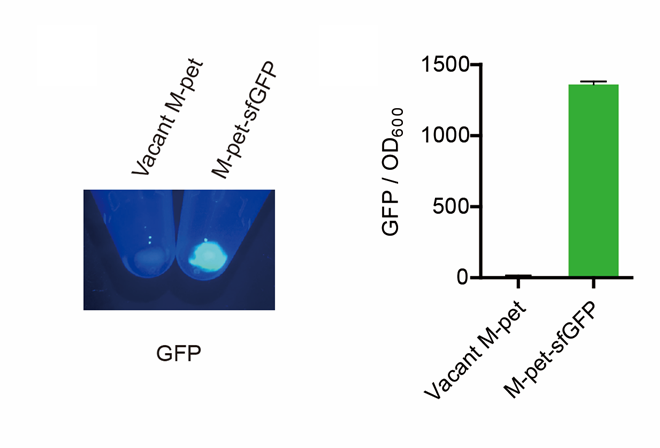
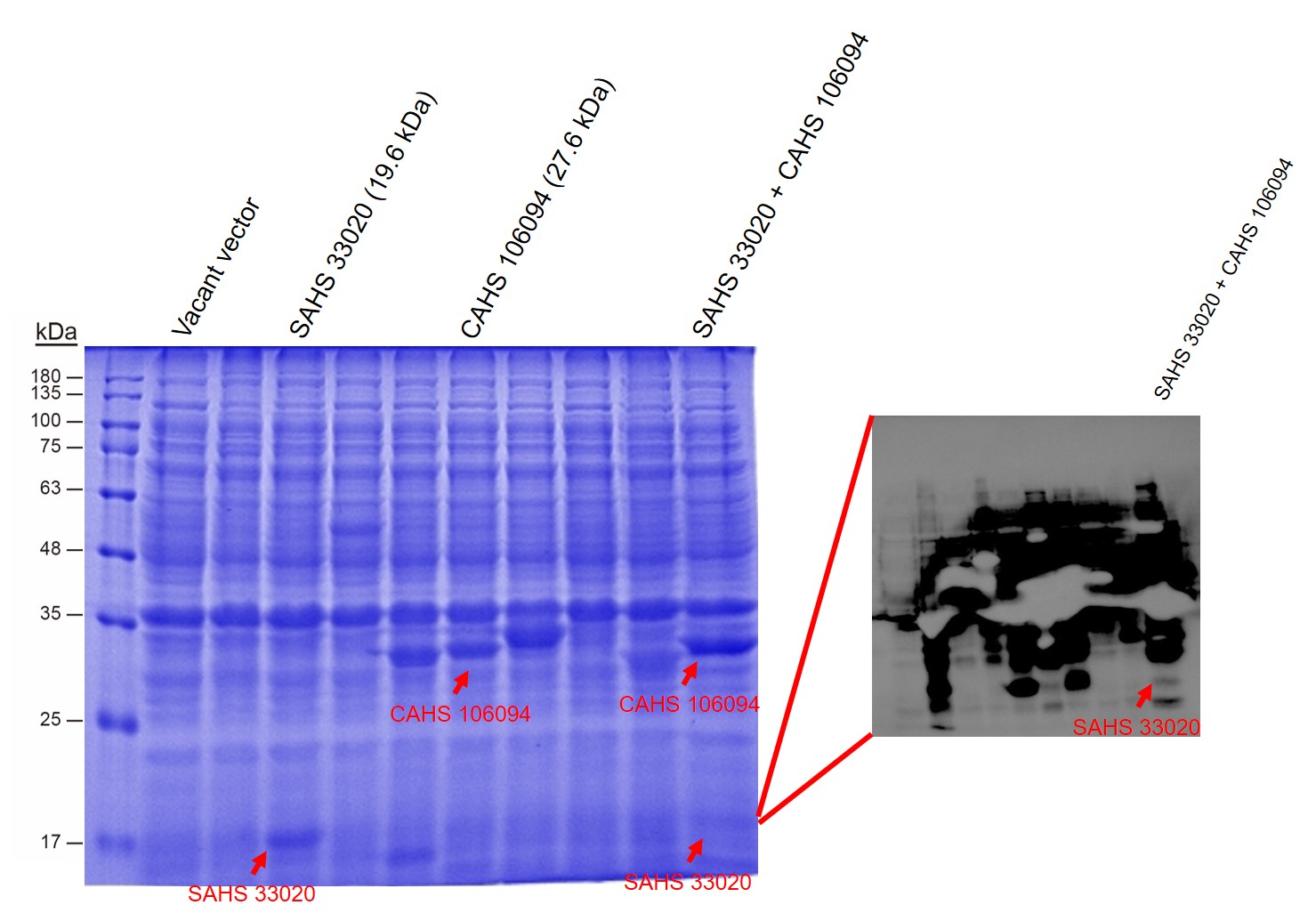
First, by a reporter, sfGFP, we confirmed that the plasmid can normally expressed exogenous proteins in E. coli BL21 strain (Fig. 5). Via SDS-PAGE, we observed the band of CAHS 106094, further verified the successfully expression (Fig.6).
Then, by freeze-drying and recover experiment, we found that compared with the control group (sfGFP), the bacteria expressing SAHS 33020 or CAHS 106094 had a better survival rate, while co-expressing the two proteins gave a better survival rate than that of expressing single TDP (Fig. 6).
The dry storage time is 2 days, which is a little short. So we waited for another 10 days to test if TDPs work in long-term storage. In accord with the result above, co-expressing two TDPs gave the best survival rate. (Fig. 6).
Summary:
CAHS 106094 helps the bacteria survive freeze-drying and subsequent dry storage. Additional SAHS 33020 can collaborate with CAHS 106094 and gave a better effect. To achieve that, you need only to transform the sequence into your bacteria.
Reference:
[1] Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., Rebecchi, L., Pielak, G.J., Koshland, D., and Goldstein, B. (2017). Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 65, 975-984 e975.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 764
Illegal NheI site found at 1759 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 27
Illegal BsaI site found at 847
Illegal BsaI.rc site found at 2
Illegal BsaI.rc site found at 822
| None |