Difference between revisions of "Part:BBa K2973021"
Nickdelkis (Talk | contribs) (→Thessaly 2019 Characterization) |
Akatsaouni (Talk | contribs) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2973021 short</partinfo> | <partinfo>BBa_K2973021 short</partinfo> | ||
| − | + | β-lactamase is an enzyme found widely in the procaryotes and confers resistance to ampicillin. This composite part comprises of the TEM-optimized β-lactamase CDS (<partinfo>BBa_I757010</partinfo>), regulated by the 105 constitutive Anderson Family Promoter (<partinfo>BBa_J23105</partinfo>) and a universal RBS (<partinfo>BBa_B0034</partinfo>). Transcription is terminated by a double terminator (<partinfo>BBa_B0015</partinfo>). This allows for enhanced protein expression in E. coli cells. | |
===Thessaly 2019 Characterization=== | ===Thessaly 2019 Characterization=== | ||
| − | Thessaly 2019 sought to <b>characterize</b> the coding sequence of <b>TEM-optimized | + | <p id="10">Thessaly 2019 sought to <b>characterize</b> the coding sequence of <b>TEM-optimized β-lactamase</b> (<partinfo>BBa_I757010</partinfo>) <b>under the regulation of the constituve Anderson Family promoters</b> <partinfo>BBa_J23100</partinfo>, <partinfo>BBa_J23105</partinfo>, <partinfo>BBa_J23106</partinfo>, <partinfo>BBa_J23119</partinfo>. β-lactamase is an enzyme that hydrolyses β-lactams (e.g. ampicillin) and is naturally found in prokaryotic cells. A colorimetric assay has been developed using nitrocefin as a substrate which after hydrolysis from β-lactamase changes the reaction color, from yellow (380nm) to red (490nm).</p> |
| − | To achieve that, the coding sequence was assembled with each promoter, a <b>universal RBS</b> ( | + | To achieve that, the coding sequence was assembled with each promoter, a <b>universal RBS</b> (<partinfo>BBa_B0034</partinfo>) and a <b>double terminator</b>(<partinfo>BBa_B0015</partinfo>). The parts were cloned in pSB1C3 and pSB1K3 and transformed into <i>E. coli DH5a</i> competent cells. |
In the photo below you can see the results of the primer addition using <b>overhang PCR</b>: | In the photo below you can see the results of the primer addition using <b>overhang PCR</b>: | ||
| Line 18: | Line 18: | ||
<body> | <body> | ||
| − | <img src="https:// | + | <img src="https://2019.igem.org/wiki/images/3/34/T--Thessaly--Contribution_gel.png" class= "center" width="300" |
| − | height=" | + | height="300"> |
<p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 1.</b> The results obtained after the PCR with the overhang primers for the different promoters of the Anderson family. We tested different annealing temperatures (45, 47 & 53℃) aiming for clear results. The expected band is at 1119bp and the ladder used was the 100bp DNA ladder by NEB.</p> | <p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 1.</b> The results obtained after the PCR with the overhang primers for the different promoters of the Anderson family. We tested different annealing temperatures (45, 47 & 53℃) aiming for clear results. The expected band is at 1119bp and the ladder used was the 100bp DNA ladder by NEB.</p> | ||
</body> | </body> | ||
| Line 28: | Line 28: | ||
| − | For the | + | For the β-lactamase assay, we set up the following experimental design: |
| − | 1. Grow BL21 (DE3) pre-culture overnight in 5ml LB (~16h) at a | + | 1. Grow BL21 (DE3) pre-culture overnight in 5ml LB (~16h) at a shaking incubator, 37℃ / 210rpm |
2. The following morning, measure the OD600 of overnight cultures | 2. The following morning, measure the OD600 of overnight cultures | ||
| Line 36: | Line 36: | ||
3. Dilute all cultures to OD600¬ = 0.05 in M9 minimal medium | 3. Dilute all cultures to OD600¬ = 0.05 in M9 minimal medium | ||
| − | 4. Grow cells 37 | + | 4. Grow cells 37℃ /210 RPM until OD600=0.4-0.6 (~2h) |
5. Dilute all cells to the same OD600 (e.g. 0.4) | 5. Dilute all cells to the same OD600 (e.g. 0.4) | ||
| Line 44: | Line 44: | ||
7. Measure the absorbance at 490nm (for nitrocefin hydrolysis) and 600nm (for cell growth) every 30 seconds for 2 hours in a microplate reader. Shake between readings. Because plateau was reached within the first 30 minutes of the reaction, only those are depicted in the graph. | 7. Measure the absorbance at 490nm (for nitrocefin hydrolysis) and 600nm (for cell growth) every 30 seconds for 2 hours in a microplate reader. Shake between readings. Because plateau was reached within the first 30 minutes of the reaction, only those are depicted in the graph. | ||
| − | To ensure that the absorbance shown corresponds only to enzymatic activity by | + | To ensure that the absorbance shown corresponds only to enzymatic activity by β-lactamase, <b>we included 3 controls in the experiment</b>. |
The first control has <b>M9 medium only</b> (no cells) and nitrocefin, the second has <b>empty BL21 (DE3) cells (no plasmid)</b> and nitrocefin, while the third has <b>BL21 (DE3) cells containing the plasmid but not the part (empty plasmid)</b>. | The first control has <b>M9 medium only</b> (no cells) and nitrocefin, the second has <b>empty BL21 (DE3) cells (no plasmid)</b> and nitrocefin, while the third has <b>BL21 (DE3) cells containing the plasmid but not the part (empty plasmid)</b>. | ||
To obtain comparable results, we normalized all values by dividing OD490 by OD600. | To obtain comparable results, we normalized all values by dividing OD490 by OD600. | ||
| Line 59: | Line 59: | ||
<img src="https://static.igem.org/mediawiki/parts/6/65/T--Thessaly--Graph-contrib.png" class= "center" width="800" | <img src="https://static.igem.org/mediawiki/parts/6/65/T--Thessaly--Graph-contrib.png" class= "center" width="800" | ||
height="508"> | height="508"> | ||
| − | <p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 2.</b> The hydrolysis of nitrocefin enabled by the expression of the | + | <p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 2.</b> The hydrolysis of nitrocefin enabled by the expression of the β-lactamase gene, under the control of different promoters (J23100, J23105, J23106 & J23119) of the Anderson family. The substrate (nitrocefin) hydrolysis (490nm) is divided by cell growth (600nm), in order to normalize all values.</p> |
</body> | </body> | ||
</html> | </html> | ||
| − | <p> The maximum expression of | + | <p> The maximum expression of β-lactamase was observed under control of the J23119 (brown line) which is the wild type promoter of the Anderson family. The expression is reduced with the J23100 and J23106 (yellow and purple line respectively), while the lowest expression levels were observed with the J23105 promoter (blue line). These results are in accordance with those from previous teams that measured fluorescence and the same pattern is observed. The controls conditions (pSB1C3 and BL21, or light purple and light blue respectively) confirm that the absorbance measured derives from β-lactamase activity only, both quantitatively and visually.</p> |
Below you can see the 96-well plate of the assay: | Below you can see the 96-well plate of the assay: | ||
| Line 75: | Line 75: | ||
<img src="https://static.igem.org/mediawiki/parts/7/7b/T--Thessaly--plate_reader_contribution.png" class= "center" width="800" | <img src="https://static.igem.org/mediawiki/parts/7/7b/T--Thessaly--plate_reader_contribution.png" class= "center" width="800" | ||
height="467"> | height="467"> | ||
| − | <p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 3.</b>The observed color change due to the hydrolyzation of nitrocefin due to the production of | + | <p style="text-align: justify; font-size: 14px; font-family: MuliLight; color: black; margin-left: auto; margin-right: auto;"><b>Figure 3.</b>The observed color change due to the hydrolyzation of nitrocefin due to the production of β-lactamase, after a 2-hour enzymatic assay.</p> |
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 13:18, 21 October 2019
TEM-optimized beta-lactamase / J23105
β-lactamase is an enzyme found widely in the procaryotes and confers resistance to ampicillin. This composite part comprises of the TEM-optimized β-lactamase CDS (BBa_I757010), regulated by the 105 constitutive Anderson Family Promoter (BBa_J23105) and a universal RBS (BBa_B0034). Transcription is terminated by a double terminator (BBa_B0015). This allows for enhanced protein expression in E. coli cells.
Thessaly 2019 Characterization
Thessaly 2019 sought to characterize the coding sequence of TEM-optimized β-lactamase (BBa_I757010) under the regulation of the constituve Anderson Family promoters BBa_J23100, BBa_J23105, BBa_J23106, BBa_J23119. β-lactamase is an enzyme that hydrolyses β-lactams (e.g. ampicillin) and is naturally found in prokaryotic cells. A colorimetric assay has been developed using nitrocefin as a substrate which after hydrolysis from β-lactamase changes the reaction color, from yellow (380nm) to red (490nm).
To achieve that, the coding sequence was assembled with each promoter, a universal RBS (BBa_B0034) and a double terminator(BBa_B0015). The parts were cloned in pSB1C3 and pSB1K3 and transformed into E. coli DH5a competent cells.
In the photo below you can see the results of the primer addition using overhang PCR:
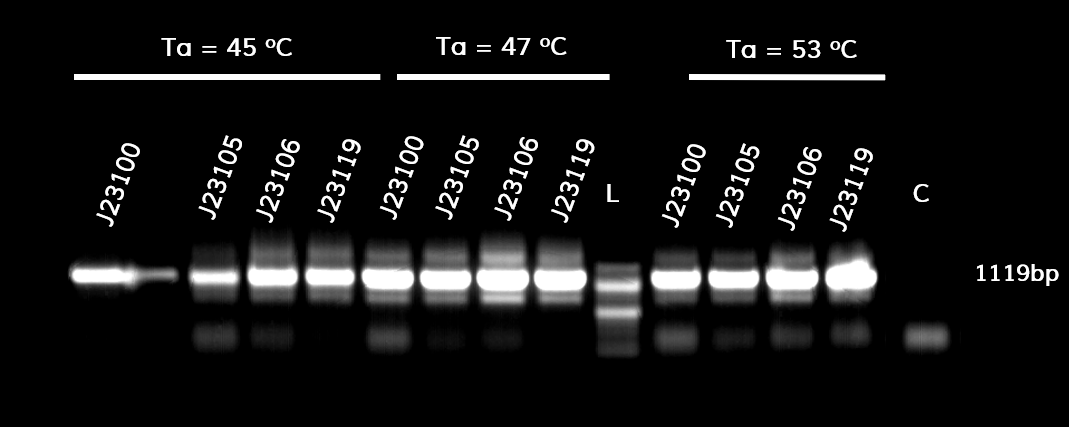
Figure 1. The results obtained after the PCR with the overhang primers for the different promoters of the Anderson family. We tested different annealing temperatures (45, 47 & 53℃) aiming for clear results. The expected band is at 1119bp and the ladder used was the 100bp DNA ladder by NEB.
For protein expression, the plasmids were transformed into E. coli BL21 (DE3) competent cells.
For the β-lactamase assay, we set up the following experimental design:
1. Grow BL21 (DE3) pre-culture overnight in 5ml LB (~16h) at a shaking incubator, 37℃ / 210rpm
2. The following morning, measure the OD600 of overnight cultures
3. Dilute all cultures to OD600¬ = 0.05 in M9 minimal medium
4. Grow cells 37℃ /210 RPM until OD600=0.4-0.6 (~2h)
5. Dilute all cells to the same OD600 (e.g. 0.4)
6. Load 160 of culture in a 96-well plate (do triplicates). Add 40 ul 0.5 uM nitrocefin for a final concentration of 100nM
7. Measure the absorbance at 490nm (for nitrocefin hydrolysis) and 600nm (for cell growth) every 30 seconds for 2 hours in a microplate reader. Shake between readings. Because plateau was reached within the first 30 minutes of the reaction, only those are depicted in the graph.
To ensure that the absorbance shown corresponds only to enzymatic activity by β-lactamase, we included 3 controls in the experiment. The first control has M9 medium only (no cells) and nitrocefin, the second has empty BL21 (DE3) cells (no plasmid) and nitrocefin, while the third has BL21 (DE3) cells containing the plasmid but not the part (empty plasmid). To obtain comparable results, we normalized all values by dividing OD490 by OD600.
The results are shown in the graph below

Figure 2. The hydrolysis of nitrocefin enabled by the expression of the β-lactamase gene, under the control of different promoters (J23100, J23105, J23106 & J23119) of the Anderson family. The substrate (nitrocefin) hydrolysis (490nm) is divided by cell growth (600nm), in order to normalize all values.
The maximum expression of β-lactamase was observed under control of the J23119 (brown line) which is the wild type promoter of the Anderson family. The expression is reduced with the J23100 and J23106 (yellow and purple line respectively), while the lowest expression levels were observed with the J23105 promoter (blue line). These results are in accordance with those from previous teams that measured fluorescence and the same pattern is observed. The controls conditions (pSB1C3 and BL21, or light purple and light blue respectively) confirm that the absorbance measured derives from β-lactamase activity only, both quantitatively and visually.
Below you can see the 96-well plate of the assay:

Figure 3.The observed color change due to the hydrolyzation of nitrocefin due to the production of β-lactamase, after a 2-hour enzymatic assay.
Note that the picture was taken after the plate-reader assay was completed and all conditions had reached a plateau, except the controls.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 163
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 783
