Difference between revisions of "Part:BBa J61101"
Carolinarp (Talk | contribs) |
|||
| (8 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
{{JCA_Arkin_RBSFamily}} | {{JCA_Arkin_RBSFamily}} | ||
| − | + | Add more about the biology of this part here | |
| + | |||
| + | |||
| + | |||
| + | >Internal Priming Screening Characterization of BBa_J61101: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer. | ||
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | Group: Valencia_UPV iGEM 2018 | ||
Author: Adrián Requena Gutiérrez, Carolina Ropero | Author: Adrián Requena Gutiérrez, Carolina Ropero | ||
<br> | <br> | ||
| − | We have adapted the part to be able to assemble transcriptional units with the Golden Gate method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure the correct spacing for the coding sequence expression when assembled in a composite part. | + | Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure the correct spacing for the coding sequence expression when assembled in a composite part. |
Characterization of the this part was performed with the transcriptional unit [https://parts.igem.org/Part:BBa_K2656104 BBa_K2656104], which was used in a comparative RBS expression experiment with composite parts [https://parts.igem.org/Part:BBa_K2656100 BBa_K265610] and [https://parts.igem.org/Part:BBa_K2656101 BBa_K2656101]. | Characterization of the this part was performed with the transcriptional unit [https://parts.igem.org/Part:BBa_K2656104 BBa_K2656104], which was used in a comparative RBS expression experiment with composite parts [https://parts.igem.org/Part:BBa_K2656100 BBa_K265610] and [https://parts.igem.org/Part:BBa_K2656101 BBa_K2656101]. | ||
They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator. | They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator. | ||
| + | |||
| + | By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS. | ||
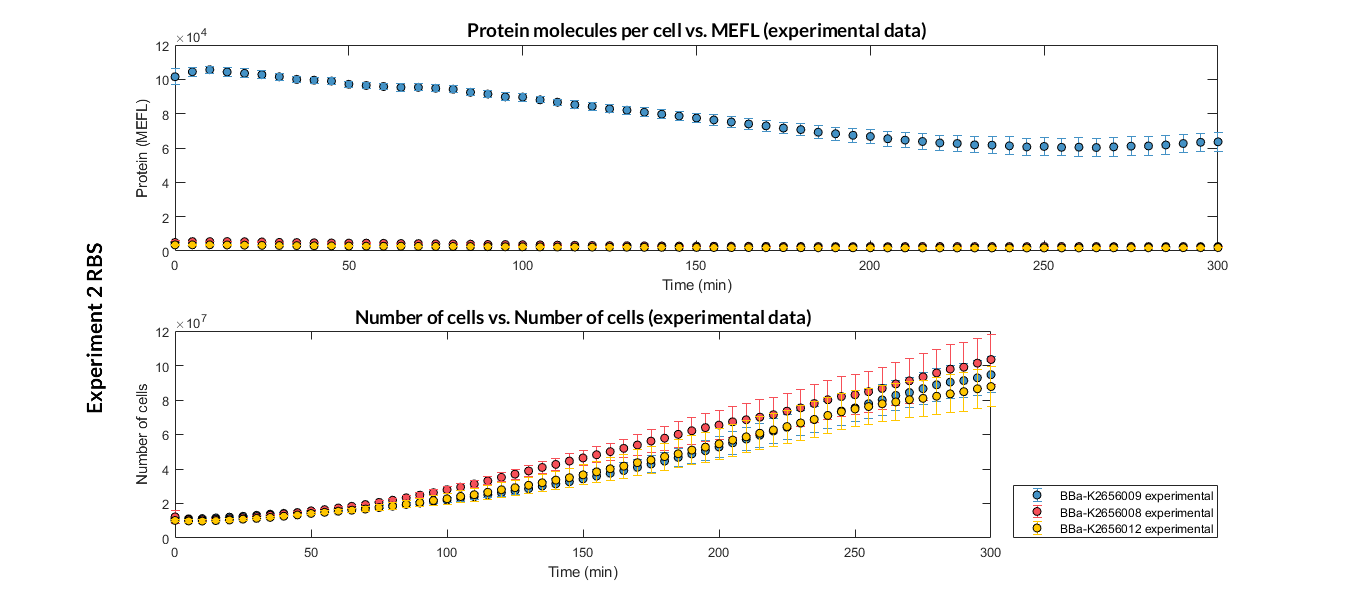
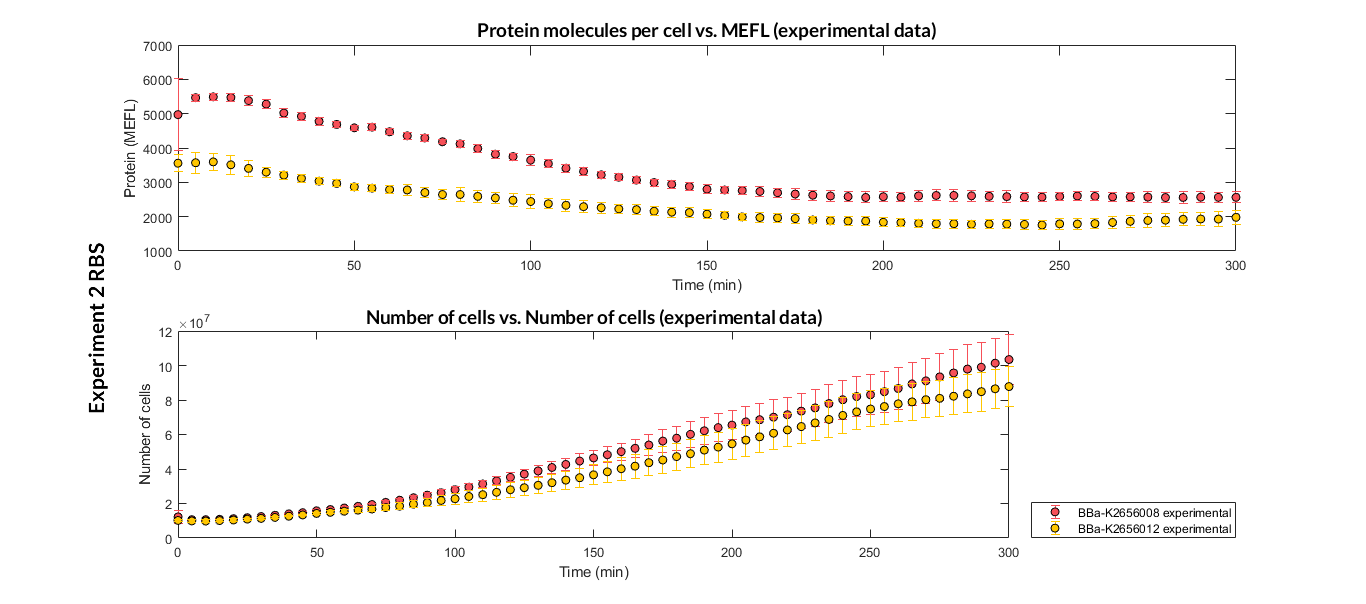
[[File:T--Valencia_UPV--optimization_exp2_RBS_all_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 1. RBS expression experiment with K2656008, K2656009 and K2656012 RBS basic pieces]] | [[File:T--Valencia_UPV--optimization_exp2_RBS_all_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 1. RBS expression experiment with K2656008, K2656009 and K2656012 RBS basic pieces]] | ||
| Line 64: | Line 73: | ||
</html> | </html> | ||
| + | <html> | ||
| + | <hr><h3 style=color:ltblue;"><a href="https://2023.igem.wiki/heidelberg/contribution">Measurements from 2023 team Heidelberg</a></h3> | ||
| + | <h4>Evaluation of RBS strength in <i>Pseudomonas fluorescens</i></h4> | ||
| + | <p>The RBS BBa_J61101 was tested in <i>P. fluorescens</i> and it's strength compared to two further ribosomal binding sites: <a href="https://parts.igem.org/Part:BBa_J61100">BBa_J61100</a> and <a href="https://parts.igem.org/Part:BBa_K4757003">BBa_K4757003</a></p> | ||
| + | <p>The parts strength was calculated by measuring the expression strength of the constitutively expressed fluorescence gene mKate2(<a href="https://parts.igem.org/Part:BBa_K4757000">BBa_K4757000</a> and calculating the fold change compared to the auto fluroescence of <i>P. fluorescens</i>.</p> | ||
| + | <img src="https://static.igem.wiki/teams/4757/wiki/fold-change-fluorescence-intensity-with-testsed-rbs.svg" alt"RBS_comp" width="300"> | ||
| + | <p display="table-caption" style="padding:10px" style="justif"><strong>Figure: Calculated fold change of fluorescence intensity of mKate2 with different ribosomal binding sites </strong><br>(RBS 1: BBa_J61100, RBS 2: BBa_J61101, RBS 3: synthetic de-novo RBS) compared to wild-type P. fluorescens. Measurements were taken in the early stationary phase. Values are presented as mean +/-SD. For statistical analysis groups were compared to wild-type P. fluorescens Reported significances were determined with ordinary One-way ANOVA with Dunn's method for multiple comparisons. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. All differences in fold changes were significant (p < 0.001 for RBS 1 and 2. To reduce complexity only edge cases are shown.</p> | ||
| + | <p> </p> | ||
| + | <p style="justify">Constructs with BBa_J61100 (RBS 1) showed minimal fold-change in fluorescence levels (0.73 +/- 0.316). The second RBS from the Anderson library (BBa_J61101, RBS 2) had a distinct increase in fold change compared to BBa_J61100 (18.15 +/- 3.21 compared to 0.73). The synthetic RBS (RBS 3) designed by the Salis-lab calculator (calculated for maximal expression strength for mKate2 mRNA) showed the strongest fluorescence (48.56 +/- 3.394).</p> | ||
| + | <p> </p> | ||
| + | </html> | ||
| − | + | ||
| − | + | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
<partinfo>BBa_J61101 parameters</partinfo> | <partinfo>BBa_J61101 parameters</partinfo> | ||
| − | < | + | <h3><center>Burden Imposed by this Part:</center></h3> |
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 3.5 ± 8.8% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 14:53, 12 October 2023
Ribosome Binding Site Family Member Parts J61100-J61150 are a family of similar ribosome binding site basic parts identified from a saturation mutagenic library.
Library TCTAGAGAAAGANNNGANNNACTAGT J61100 tctagaGAAAGAGGGGACAAactagt J61101 tctagaGAAAGACAGGACCCactagt J61102 tctagaGAAAGATCCGATGTactagt J61103 tctagaGAAAGATTAGACAAactagt J61104 tctagaGAAAGAAGGGACAGactagt J61105 tctagaGAAAGACATGACGTactagt J61106 tctagaGAAAGATAGGAGACactagt J61107 tctagaGAAAGAAGAGACTCactagt J61108 tctagaGAAAGACGAGATATactagt J61109 tctagaGAAAGACTGGAGACactagt J61110 tctagaGAAAGAGGCGAATTactagt J61111 tctagaGAAAGAGGCGATACactagt J61112 tctagaGAAAGAGGTGACATactagt J61113 tctagaGAAAGAGTGGAAAAactagt J61114 tctagaGAAAGATGAGAAGAactagt J61115 tctagaGAAAGAAGGGATACactagt J61116 tctagaGAAAGACATGAGGCactagt J61117 tctagaGAAAGACATGAGTTactagt J61118 tctagaGAAAGAGACGAATCactagt J61119 tctagaGAAAGATTTGATATactagt J61120 tctagaGAAAGACGCGAGAAactagt J61121 tctagaGAAAGAGACGAGTCactagt J61122 tctagaGAAAGAGAGGAGCCactagt J61123 tctagaGAAAGAGATGACTAactagt J61124 tctagaGAAAGAGCCGACATactagt J61125 tctagaGAAAGAGCCGAGTTactagt J61126 tctagaGAAAGAGGTGACTCactagt J61127 tctagaGAAAGAGTGGAACTactagt J61128 tctagaGAAAGATAGGACTCactagt J61129 tctagaGAAAGATTGGACGTactagt J61130 tctagaGAAAGAAACGACATactagt J61131 tctagaGAAAGAACCGAATTactagt J61132 tctagaGAAAGACAGGATTAactagt J61133 tctagaGAAAGACCCGAGACactagt J61134 tctagaGAAAGACCGGAAATactagt J61135 tctagaGAAAGACCGGAGACactagt J61136 tctagaGAAAGAGCTGAGCAactagt J61137 tctagaGAAAGAGTAGATCAactagt J61138 tctagaGAAAGATATGAATAactagt J61139 tctagaGAAAGATTAGAGTCactagt
These parts are present in plasmid pSB1A2, but there is also a constitutive promoter (J23100-derived) inserted into the XbaI site. So, for example, the EcoRI/PstI region of part J61100 reads:
Biobrick 5' XbaI J23100 XbaI RBS Part Biobrick 3' gaattcgcggccgcttctagaGTTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGCTtctagaGAAAGAGGGGACAAactagtagcggccgctgcag
This feature in no way prevents the use of these parts in standard Biobrick assembly. Normal prefix insertion into EcoRI/XbaI will delete this promoter element. Suffix insertion into SpeI/PstI will retain this promoter, but it can of course be removed later by a prefix insertion.
Note also that the base 5' to the SpeI site is allowed to float in these parts and is therefore rarely "T". The "G" downstream of the XbaI site obeys the standard. Because the database does not permit variation at this position, the predicted sequences of composite parts derived from these parts will be incorrect at this position.
More on this family of parts and their quantitative behavior is described here.
Add more about the biology of this part here
>Internal Priming Screening Characterization of BBa_J61101: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Usage and Biology
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure the correct spacing for the coding sequence expression when assembled in a composite part.
Characterization of the this part was performed with the transcriptional unit BBa_K2656104, which was used in a comparative RBS expression experiment with composite parts BBa_K265610 and BBa_K2656101. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS.
| Table 1. Optimized parameters for the BBa_K2656008 RBS. | |||
| Parameter | Value | ||
| Translation rate p | p = 0.02889 min-1 | ||
| Dilution rate μ | μ = 0.0118 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. It has been defined as the quotient between the values of the protein in equilibrium of the results of the simulation of one RBS and the reference RBS. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.032 | ||
| p parameter ratio (pRBS/pref) | 0.032 | ||
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team Warsaw 2010's measurement
RBS strength (relative to B0034): 22,70%
Team TU Delft 2010's measurements
RBS strength: 11.9%, standard deviation: 2.14%RBS strength is relative to B0034, obtained from an average of 12 measurements. Protein production rate is calculated using our production model
Measurements from 2023 team Heidelberg
Evaluation of RBS strength in Pseudomonas fluorescens
The RBS BBa_J61101 was tested in P. fluorescens and it's strength compared to two further ribosomal binding sites: BBa_J61100 and BBa_K4757003
The parts strength was calculated by measuring the expression strength of the constitutively expressed fluorescence gene mKate2(BBa_K4757000 and calculating the fold change compared to the auto fluroescence of P. fluorescens.

Figure: Calculated fold change of fluorescence intensity of mKate2 with different ribosomal binding sites
(RBS 1: BBa_J61100, RBS 2: BBa_J61101, RBS 3: synthetic de-novo RBS) compared to wild-type P. fluorescens. Measurements were taken in the early stationary phase. Values are presented as mean +/-SD. For statistical analysis groups were compared to wild-type P. fluorescens Reported significances were determined with ordinary One-way ANOVA with Dunn's method for multiple comparisons. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. All differences in fold changes were significant (p < 0.001 for RBS 1 and 2. To reduce complexity only edge cases are shown.
Constructs with BBa_J61100 (RBS 1) showed minimal fold-change in fluorescence levels (0.73 +/- 0.316). The second RBS from the Anderson library (BBa_J61101, RBS 2) had a distinct increase in fold change compared to BBa_J61100 (18.15 +/- 3.21 compared to 0.73). The synthetic RBS (RBS 3) designed by the Salis-lab calculator (calculated for maximal expression strength for mKate2 mRNA) showed the strongest fluorescence (48.56 +/- 3.394).
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.