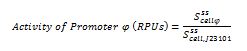Difference between revisions of "Part:BBa J23111:Experience"
| Line 42: | Line 42: | ||
| + | |} | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_R0062 AddReview 4.1691795684</partinfo> | ||
| + | <I>CLSB-UK 2017</I> | ||
| + | |width='60%' valign='top'| | ||
| + | We used part BBa_J23111 as the promoter for our six constructs that contained our toehold switches. We chose BBa_J23111 to obtain high concentrations of mRNA for studying the effect of our toehold switches on translation. We had the whole promoter-toehold switch-GFP/Luciferase constructs synthesised as gBlocks and inserted these sequences into the pSB1C3 plasmid backbone. We then attempted to amplify these plasmids in E.coli. To check that our inserts had been cloned into vector successfully, we performed a test restriction digest and ran the products on a gel. The insert and vector lengths were the correct sizes. However, upon sequencing our inserts, we found that none of the sequences corresponded to anything we were expecting to see. We used BLAST search to identify our construct sequences and found parts of GFP and Luciferase, but it appeared that at least some of the inserts had been excised from the plasmid. We therefore postulate that our constructs were toxic to the E.coli. Since minimal levels of translation should have occurred due to the presence of the toehold switches, and both GFP and Luciferase have both been expressed in E. coli without any issues, we posit that the transcript RNA is damaging to the cells. Furthermore, only the cells that survived transformations had modified BBa_J23111 sequences. Indeed, none of the sequences examined had any part of the promoter, thus transcription of the constructs had been inactivated. This data therefore indicates that BBa_J23111 functions as a constitutive promoter. We also believe that the promoter is reasonably strong as it produced toxic levels of our transcript. We therefore recommend that this promoter should therefore not be used for expressing constructs that have not already been characterised in cells. | ||
|} | |} | ||
Latest revision as of 18:57, 20 September 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J23111
User Reviews
UNIQce01bad4aac7bdd1-partinfo-00000000-QINU
|
•••••
ETH Zurich 2014 |
Characterization of the pLux promoter's sensitivity to 3OC6-HSL depending on LuxR under the promoters J23100, J23111, and J23109The amount of regulator LuxR (BBa_C0062) in the system was shown to influence the pLux promoter's response to the inducer concentration (3OC6-HSL). By using the three different constitutive promoters BBa_J23100, BBa_J23109, and BBa_J23111 for the production of LuxR we have measured this effect in terms of fluorescence/OD600 (see Figure 1). Background informationWe used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter pLux (BBa_R0062) followed by RBS (BBa_B0034) and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts BBa_B0040 and BBa_B0015 were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and one of three promoters chosen from the Anderson promoter collection followed by luxR (BBa_C0062). The detailed regulator construct design and full sequences (piG0041, piG0046, piG0047) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. Experimental Set-UpThe above described E. coli TOP10 strains were grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD600 of about 1.5 (37 °C, 220 rpm). As a reference, a preculture of the same strain lacking the sfGFP gene was included for each assay. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in triplicates in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h at 37 °C with a Tecan infinite M200 PRO plate reader (optical density measured at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). After 200 min we added the following concentrations of inducers (3OC6-HSL, 3OC12-HSL, and C4-HSL): 10-4 nM and 104 nM (from 100 mM stocks in DMSO). Attention: All the dilutions of 3OC12-HSL should be made in DMSO to avoid precipitation. In addition, in one triplicate only H2O was added as a control. From the the obtained kinetic data, we calculated mean values and plotted the dose-response-curve for 200 min past induction (see Figure 1). ResultsThe measurements of the induced system with 3OC6-HSL concentrations of 10-13 M to 10-5 M showed an increasing sensitivity of the pLux (BBa_R0062) promoter (in terms of fluorescence per OD600) for increasing strength of the promoter controlling LuxR (BBa_C0062) expression (see Figure 1). For BBa_J23100 (strongest promoter chosen) the sensitivity is highest (half maximal effective concentration EC50 approximately 20 pM), for BBa_J23109 (weakest one chosen) the sensitivity is lowest (EC50 approximately 100 pM), with BBa_J23111 (medium) falling between these two but closer to the strong promoter (EC50 approximately 10 nM). Overall, this is in line with the promoter strength given in the Anderson collection.
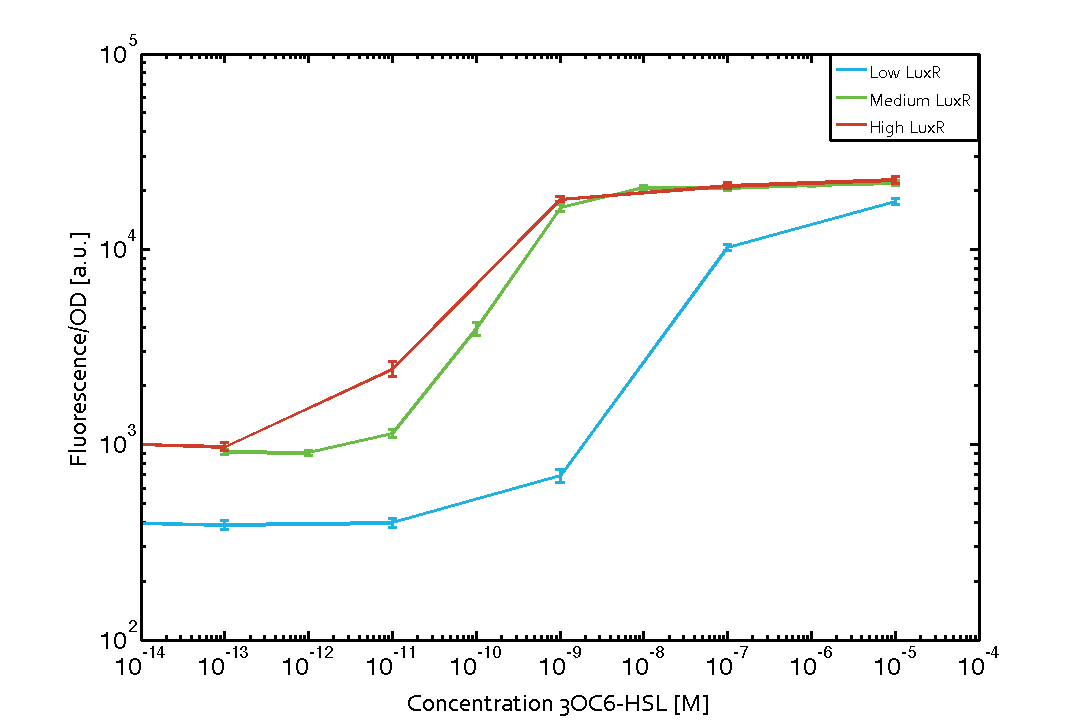 Figure 1 Influence of varied promoter strength on the expression of LuxR (BBa_C0062) and the resulting effect on pLux (BBa_R0062). The fluorescence per OD600 is shown over an inducer-range of 10-13 M to 10-5 M. The promoters used are: BBa_J23100 (high LuxR expression, red), BBa_J23111 (medium LuxR expression, green), and BBa_J23109 (low LuxR expression, blue). All three promoters are part of the Anderson collection. Data points are mean values of triplicate measurements in 96-well microtiter plates 200 min after induction ± standard deviation. For the full data set and kinetics please [http://2014.igem.org/Team:ETH_Zurich/contact contact] us or visit the [http://2014.igem.org/Team:ETH_Zurich/data/raw raw data] page.
|
|
••••
CLSB-UK 2017 |
We used part BBa_J23111 as the promoter for our six constructs that contained our toehold switches. We chose BBa_J23111 to obtain high concentrations of mRNA for studying the effect of our toehold switches on translation. We had the whole promoter-toehold switch-GFP/Luciferase constructs synthesised as gBlocks and inserted these sequences into the pSB1C3 plasmid backbone. We then attempted to amplify these plasmids in E.coli. To check that our inserts had been cloned into vector successfully, we performed a test restriction digest and ran the products on a gel. The insert and vector lengths were the correct sizes. However, upon sequencing our inserts, we found that none of the sequences corresponded to anything we were expecting to see. We used BLAST search to identify our construct sequences and found parts of GFP and Luciferase, but it appeared that at least some of the inserts had been excised from the plasmid. We therefore postulate that our constructs were toxic to the E.coli. Since minimal levels of translation should have occurred due to the presence of the toehold switches, and both GFP and Luciferase have both been expressed in E. coli without any issues, we posit that the transcript RNA is damaging to the cells. Furthermore, only the cells that survived transformations had modified BBa_J23111 sequences. Indeed, none of the sequences examined had any part of the promoter, thus transcription of the constructs had been inactivated. This data therefore indicates that BBa_J23111 functions as a constitutive promoter. We also believe that the promoter is reasonably strong as it produced toxic levels of our transcript. We therefore recommend that this promoter should therefore not be used for expressing constructs that have not already been characterised in cells. |
iGEM CINVESTAV_IPN_UNAM CHARACTERIZATION OF IGEM DISTRIBUTION BIOPARTS
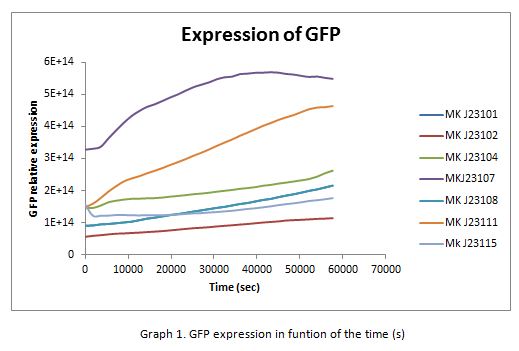
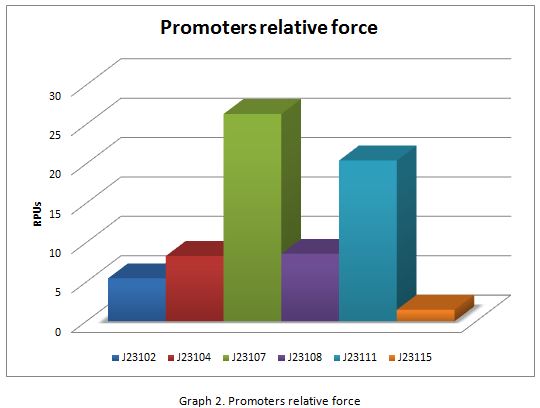
For contribute to the parts registry our team decided to make the characterization of constitutive promoters, in E. coli, belonging to the family isolated from a small combinatorial library (J23101 , J23102, J23104, J23107, J23108, J2311, and J23115) which were attached to GFP, in psB1C3, to determine promoter activity, using the equipment Victor X3 Multilabel Plate Reader.
Fig. 1 Construction of the promoter J23111 expressing GFP.
Methods
With the selected colonies, an overnight culture was made in M9 media(minimal media supplemented with 0.2% CAA). After 12 hours the culture was transferred to a 96 well plate at a 1:10 dilution (20 μl of culture and 180 μL of fresh M9 medium). OD and fluorescence measurements of the selected colonies were performed at intervals of 30 minutes for 16 h. From the results the PopS were calculated (polymerases per second).
Modeling
The ecuations used for calulated de promoter activity were based on (R. K. Jason et. al 2009).
Results
In the following graphs there is shown the GFP expression in function of th time and the realtive promotor intensity.
With the previous results of the characterization of the promoters there is concluded that the promoter J23107, is the strongest because it produces more RPUs”