Difference between revisions of "Part:BBa K731722"
| (19 intermediate revisions by 3 users not shown) | |||
| Line 10: | Line 10: | ||
Since many different teams had problems using [[Part:BBa_B0010|BBa_B0010]], this is the same part newly submitted, confirmed by sequencing and by experience...ready to use! | Since many different teams had problems using [[Part:BBa_B0010|BBa_B0010]], this is the same part newly submitted, confirmed by sequencing and by experience...ready to use! | ||
| − | The characterization of this | + | The characterization of this part was done by the Trento iGEM team 2012 using the new platforms for terminator characterization that they have built and submitted to the Registry as [[Part:BBa_K731700|BBa_K731700]] and [[Part:BBa_K731710|BBa_K731710]]. |
| − | + | ||
|} | |} | ||
| Line 19: | Line 18: | ||
| − | + | <!-- --> | |
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K731722 SequenceAndFeatures</partinfo> | ||
<hr> | <hr> | ||
| − | ''' | + | '''Usage and Biology''' |
| − | |||
| − | + | We made some preliminary analyses to define the best procedure to characterize this terminator using [[Part:BBa_K731700|BBa_K731700]] and [[Part:BBa_K731710|BBa_K731710]]. We found that the emission from A206K Venus (i.e. mVenus) was much stronger than the mCherry emission, resulting in some spectral overlap. To improve the quality of the data, off-peak excitation and emission wavelengths were identified that minimized the effect. Therefore, mVenus was excitated at 485 nm (excitation peak is at 515 nm), and the emission of mVenus was collected at its maximum position, i.e. 528 nm. The maximum mCherry excitation wavelength was used (587 nm), but the emission of mCherry was read at 615 nm, as opposed to the maximum at 609 nm. Here is a summary of these wavelengths: | |
| + | <div style="text-align:center"> | ||
| + | |||
| + | {|border = "1" cellpadding="5" cellspacing="0" align="centre" | ||
| + | | | ||
| + | |Standard Excitation (nm) | ||
| + | |Standard Emission (nm) | ||
| + | |Modified Excitation (nm) | ||
| + | |Modified Emission (nm) | ||
| + | |- | ||
| + | |mCherry | ||
| + | |587 | ||
| + | |609 | ||
| + | |587 | ||
| + | |615 | ||
| + | |- | ||
| + | |mVenus | ||
| + | |515 | ||
| + | |528 | ||
| + | |485 | ||
| + | |528 | ||
| + | |} | ||
| + | </div> | ||
| + | <html> | ||
| + | <p style="width:900px; text-align:justify "><em><strong>TABLE 1.</strong> <b>Standard and modified excitation and emission wavelengths </b> <br/> | ||
| + | </em> </p> </html> | ||
| + | |||
| + | |||
| + | The activity of this part was analyzed both with T7 and ''E. coli'' RNA polymerases ''in vivo'' and with T7 RNA polymerase ''in vitro''. | ||
| + | The activity of this terminator was measured as the ratio between the two proteins' levels using the equation found in literature for termination efficiency. | ||
| + | Doing this measurements, however, we realized that terminators can have an effect also on mCherry expression, enhancing it. This effect was previously described <cite>Abe</cite>. As a consequence of this unexpected outcome, we defined other two parameters that are here summarized in addition to the literature definition of termination efficiency. | ||
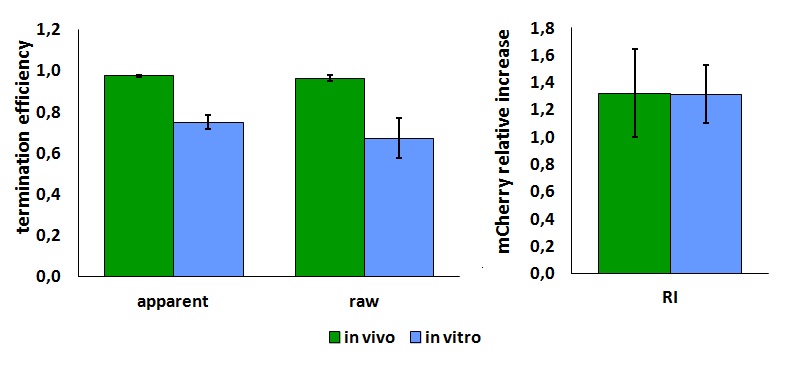
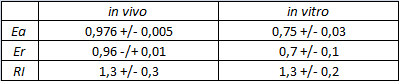
The parameters used to analyze the data are: | The parameters used to analyze the data are: | ||
| − | apparent termination efficiency, [[Image:FormulaEa.jpg]] | + | apparent termination efficiency, calculated with the equation found in literature <cite>Nojima</cite>, [[Image:FormulaEa.jpg]] |
| − | raw termination efficiency, [[Image:FormulaEr.jpg]] | + | raw termination efficiency, that does not consider the mCherry contribution [[Image:FormulaEr.jpg]] |
relative increase in the upstream gene expression, [[Image:RIequation.png]] | relative increase in the upstream gene expression, [[Image:RIequation.png]] | ||
| − | |||
where | where | ||
| − | -Vs is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker | + | -''Vs'' is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker |
| − | -Vc is the A206K Venus peak’s intensity of the control construct | + | -''Vc'' is the A206K Venus peak’s intensity of the control construct without intervening terminator |
| − | -Cs is the mCherry peak’s intensity of the construct with the terminator inserted | + | -''Cs'' is the mCherry peak’s intensity of the construct with the terminator inserted |
| − | -Cc is the mCherry peak’s intensity of the control construct | + | -''Cc'' is the mCherry peak’s intensity of the control construct |
| + | '''''In vivo'' measurements:''' | ||
| − | ''' | + | Our experiments exploited an ''E. coli'' lysogen strain carrying T7 RNA polymerase and lacIq. Additionally, the cells, i.e. ''E. coli'' BL21(DE3) pLysS, also contained a plasmid encoding T7 lysozyme and chloramphenicol resistance. T7 lysozyme is a natural inhibitor of T7 RNA polymerase activity, thus reducing background expression of the target genes. The T7 RNA polymerase is behind a lacUV5 promoter. |
<div style="text-align:center">[[Image:Ecoliterminatorinvivo.jpg]]</div> | <div style="text-align:center">[[Image:Ecoliterminatorinvivo.jpg]]</div> | ||
| Line 58: | Line 88: | ||
<div style="text-align:center">[[Image:Ecoliterminatortabinvivo.jpg]]</div> | <div style="text-align:center">[[Image:Ecoliterminatortabinvivo.jpg]]</div> | ||
<p style="width:600px; margin-left:150px; margin-bottom:60px; | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
| − | text-align:justify "><em><strong>FIGURE 1.</strong> '''E.coli terminator's effect on protein expression with two different RNA polymerases'''<br/>The data shown in figure 1 were acquired in two different days. For each day 4 different replicates were measured at different times. | + | text-align:justify "><em><strong>FIGURE 1.</strong> '''''E.coli'' terminator's effect on protein expression with two different RNA polymerases'''<br/>The data shown in figure 1 were acquired in two different days. For each day 4 different replicates were measured at different times. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Briefly, BL21(DE3)pLysS were grown in 10 mL of LB until an OD of 0.6 was reached and induced with 0.5 mM IPTG. After 3 hours of induction, 4 separate aliquots of 1 mL were taken and sonicated 3 times for 10 seconds at intervals of 30 seconds. After sonication the samples were diluted 1:2 with PBS 1X directly into a cuvette and incubated overnight at 4°C. Fluorescence measurements were taken with a Cary Eclipse Varian fluorimeter with a window ranging from 450 nm to 700 nm, a slit of 5nm, a voltage of 570V for the characterization with T7 polymerase and of 520V for the measurements with ''E. coli'' RNA polymerase. | ||
| + | </em></p> | ||
'''In vitro measurements:''' | '''In vitro measurements:''' | ||
| + | |||
| + | We performed the in vitro analysis only with the T7 RNA polymerase. | ||
<div style="text-align:center">[[Image:Ecoliterminatorinvitro.jpg]]</div> | <div style="text-align:center">[[Image:Ecoliterminatorinvitro.jpg]]</div> | ||
| Line 72: | Line 102: | ||
<div style="text-align:center">[[Image:Ecoliterminatortabinvitro.jpg]]</div> | <div style="text-align:center">[[Image:Ecoliterminatortabinvitro.jpg]]</div> | ||
<p style="width:600px; margin-left:150px; margin-bottom:60px; | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
| − | text-align:justify "><em><strong>FIGURE 2.</strong> '''E.coli terminator's effect on in vitro protein synthesis with the T7 RNA polymerases'''<br/>Cell free measurements were done with the PurExpress kit from New England Biolabs, using 250 ng of DNA previously purified by ethanol precipitation, following the protocol suggested by the manufacturer. Measurements were done with a PTI fluorimeter | + | text-align:justify "><em><strong>FIGURE 2.</strong> '''E.coli terminator's effect on in vitro protein synthesis with the T7 RNA polymerases'''<br/>Cell free measurements were done with the PurExpress kit from New England Biolabs, using 250 ng of DNA previously purified by ethanol precipitation, following the protocol suggested by the manufacturer. Measurements were done with a PTI fluorimeter. |
</em> </p> | </em> </p> | ||
| + | We submitted also this Part inside both [[Part:BBa_K731700|BBa_K731700]] (T7 promoter backbone) and [[Part:BBa_K731710|BBa_K731710]] (''tac'' promoter backbone) as, respecitively, [[Part:BBa_K731701|BBa_K731701]] and [[Part:BBa_K731702|BBa_K731702]]. We hope that this could be helpful for anyone who want to verify and further delve into our results. | ||
| + | |||
| + | More information about the procedure used, the results obtained and our considerations can be found in the [http://2012.igem.org/Team:UNITN-Trento| Trento iGEM 2012 wiki page]. | ||
| + | |||
| + | ==References== | ||
| + | <biblio> | ||
| + | #Nojima pmid=16379390 | ||
| + | #Abe pmid=9150882 | ||
| + | </biblio> | ||
| + | |||
| + | |||
| − | + | ==Functional Parameters: Austin_UTexas== | |
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_K731722 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: -2.7 ± 3.6% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 09:51, 13 June 2021
|
T1 terminator from E. coli rrnB
Since many different teams had problems using BBa_B0010, this is the same part newly submitted, confirmed by sequencing and by experience...ready to use! The characterization of this part was done by the Trento iGEM team 2012 using the new platforms for terminator characterization that they have built and submitted to the Registry as BBa_K731700 and BBa_K731710. |
Examples of Secondary Structures
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
We made some preliminary analyses to define the best procedure to characterize this terminator using BBa_K731700 and BBa_K731710. We found that the emission from A206K Venus (i.e. mVenus) was much stronger than the mCherry emission, resulting in some spectral overlap. To improve the quality of the data, off-peak excitation and emission wavelengths were identified that minimized the effect. Therefore, mVenus was excitated at 485 nm (excitation peak is at 515 nm), and the emission of mVenus was collected at its maximum position, i.e. 528 nm. The maximum mCherry excitation wavelength was used (587 nm), but the emission of mCherry was read at 615 nm, as opposed to the maximum at 609 nm. Here is a summary of these wavelengths:
| Standard Excitation (nm) | Standard Emission (nm) | Modified Excitation (nm) | Modified Emission (nm) | |
| mCherry | 587 | 609 | 587 | 615 |
| mVenus | 515 | 528 | 485 | 528 |
TABLE 1. Standard and modified excitation and emission wavelengths
The activity of this part was analyzed both with T7 and E. coli RNA polymerases in vivo and with T7 RNA polymerase in vitro.
The activity of this terminator was measured as the ratio between the two proteins' levels using the equation found in literature for termination efficiency.
Doing this measurements, however, we realized that terminators can have an effect also on mCherry expression, enhancing it. This effect was previously described Abe. As a consequence of this unexpected outcome, we defined other two parameters that are here summarized in addition to the literature definition of termination efficiency.
The parameters used to analyze the data are:
apparent termination efficiency, calculated with the equation found in literature Nojima, 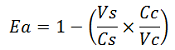
raw termination efficiency, that does not consider the mCherry contribution 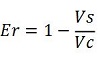
relative increase in the upstream gene expression, 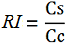
where
-Vs is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker
-Vc is the A206K Venus peak’s intensity of the control construct without intervening terminator
-Cs is the mCherry peak’s intensity of the construct with the terminator inserted
-Cc is the mCherry peak’s intensity of the control construct
In vivo measurements:
Our experiments exploited an E. coli lysogen strain carrying T7 RNA polymerase and lacIq. Additionally, the cells, i.e. E. coli BL21(DE3) pLysS, also contained a plasmid encoding T7 lysozyme and chloramphenicol resistance. T7 lysozyme is a natural inhibitor of T7 RNA polymerase activity, thus reducing background expression of the target genes. The T7 RNA polymerase is behind a lacUV5 promoter.
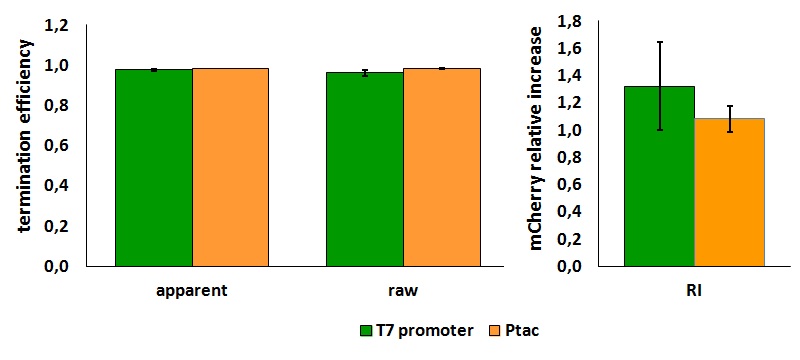
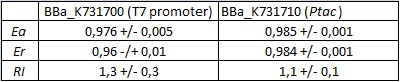
FIGURE 1. E.coli terminator's effect on protein expression with two different RNA polymerases
The data shown in figure 1 were acquired in two different days. For each day 4 different replicates were measured at different times.
Briefly, BL21(DE3)pLysS were grown in 10 mL of LB until an OD of 0.6 was reached and induced with 0.5 mM IPTG. After 3 hours of induction, 4 separate aliquots of 1 mL were taken and sonicated 3 times for 10 seconds at intervals of 30 seconds. After sonication the samples were diluted 1:2 with PBS 1X directly into a cuvette and incubated overnight at 4°C. Fluorescence measurements were taken with a Cary Eclipse Varian fluorimeter with a window ranging from 450 nm to 700 nm, a slit of 5nm, a voltage of 570V for the characterization with T7 polymerase and of 520V for the measurements with E. coli RNA polymerase.
In vitro measurements:
We performed the in vitro analysis only with the T7 RNA polymerase.
FIGURE 2. E.coli terminator's effect on in vitro protein synthesis with the T7 RNA polymerases
Cell free measurements were done with the PurExpress kit from New England Biolabs, using 250 ng of DNA previously purified by ethanol precipitation, following the protocol suggested by the manufacturer. Measurements were done with a PTI fluorimeter.
We submitted also this Part inside both BBa_K731700 (T7 promoter backbone) and BBa_K731710 (tac promoter backbone) as, respecitively, BBa_K731701 and BBa_K731702. We hope that this could be helpful for anyone who want to verify and further delve into our results.
More information about the procedure used, the results obtained and our considerations can be found in the [http://2012.igem.org/Team:UNITN-Trento| Trento iGEM 2012 wiki page].
References
<biblio>
- Nojima pmid=16379390
- Abe pmid=9150882
</biblio>
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.