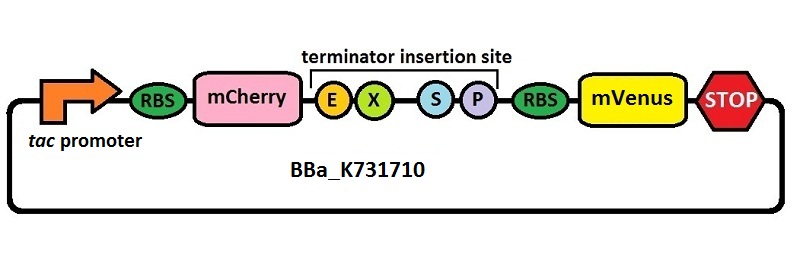
Difference between revisions of "Part:BBa K731710"
| (8 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K731710 short</partinfo> | <partinfo>BBa_K731710 short</partinfo> | ||
| − | This is a platform for the characterization of terminator efficiency by fluorimetric measurements and ratiometric analysis under IPTG inducible tac promoter’s control | + | This is a platform for the characterization of terminator efficiency by fluorimetric measurements and ratiometric analysis under IPTG inducible ''tac'' promoter’s control. It was built starting from [[Part:BBa_K731700|BBa_K731700]], where the T7 promoter was replaced with the ''tac'' promoter by insertion/deletion mutagenesis. The platform has been used for the analysis of T7 wild type terminator's ([[Part:BBa_K731721|BBa_K731721]]) and an ''E. coli'' terminator's ([[Part:BBa_K731722|BBa_K731722]]) effects on protein synthesis. |
| − | The combined use of BBa_K731710 and [[Part:BBa_K731700|BBa_K731700]] | + | The combined use of BBa_K731710 and [[Part:BBa_K731700|BBa_K731700]] allows to analyze also any potential difference in terminators' activity due to different RNA polymerases. |
| − | <div style="text-align:center">[[Image: | + | <div style="text-align:center">[[Image:K731710imageproject.jpg]]</div> |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
We made some preliminary analyses to define the best procedure to use this platform. | We made some preliminary analyses to define the best procedure to use this platform. | ||
We found that the emission from mVenus was much stronger than the mCherry emission, resulting in some spectral overlap. To improve the quality of the data, off-peak excitation and emission wavelengths were identified that minimized the effect. Therefore, mVenus was excitated at 485 nm (excitation peak is at 515 nm), and the emission of mVenus was collected at its maximum position, i.e. 528 nm. The maximum mCherry excitation wavelength was used (587 nm), but the emission of mCherry was read at 615 nm, as opposed to the maximum at 609 nm. | We found that the emission from mVenus was much stronger than the mCherry emission, resulting in some spectral overlap. To improve the quality of the data, off-peak excitation and emission wavelengths were identified that minimized the effect. Therefore, mVenus was excitated at 485 nm (excitation peak is at 515 nm), and the emission of mVenus was collected at its maximum position, i.e. 528 nm. The maximum mCherry excitation wavelength was used (587 nm), but the emission of mCherry was read at 615 nm, as opposed to the maximum at 609 nm. | ||
| − | Here is a summary these wavelengths: | + | Here is a summary of these wavelengths: |
| + | |||
<div style="text-align:center"> | <div style="text-align:center"> | ||
| + | |||
{|border = "1" cellpadding="5" cellspacing="0" align="centre" | {|border = "1" cellpadding="5" cellspacing="0" align="centre" | ||
| | | | ||
| − | |Excitation (nm) | + | |Standard Excitation (nm) |
| − | |Emission(nm) | + | |Standard Emission (nm) |
| + | |Modified Excitation (nm) | ||
| + | |Modified Emission (nm) | ||
|- | |- | ||
|mCherry | |mCherry | ||
|587 | |587 | ||
|609 | |609 | ||
| + | |587 | ||
| + | |615 | ||
|- | |- | ||
| − | | | + | |mVenus |
|515 | |515 | ||
|528 | |528 | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|485 | |485 | ||
|528 | |528 | ||
| − | |}</div> | + | |} |
| + | </div><html><p style="width:900px; text-align:justify "><em><strong>TABLE 1.</strong> <b>Standard and modified excitation and emission wavelengths. </b><br/></em> </p> </html> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
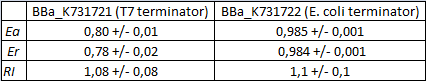
| − | This | + | This part was used to characterize a T7 wild type terminator ([[Part:BBa_K731721|BBa_K731721]]) and an ''E. coli'' terminator ([[Part:BBa_K731722|BBa_K731722]]). |
These terminators's activity was measured as the ratio between the two proteins' levels using the equation found in literature for termination efficiency. | These terminators's activity was measured as the ratio between the two proteins' levels using the equation found in literature for termination efficiency. | ||
| − | Doing this measurements, however, we realized that terminators can have an effect also on mCherry expression, enhancing it. | + | Doing this measurements, however, we realized that terminators can have an effect also on mCherry expression, enhancing it. This effect was previously described <cite>Abe</cite>. As a consequence of this unexpected outcome, we defined other two parameters that are here summarized in addition to the literature definition of termination efficiency. |
The parameters used to analyze the data are: | The parameters used to analyze the data are: | ||
| − | apparent termination efficiency | + | apparent termination efficiency, calculated with the equation found in literature <cite>Nojima</cite>, [[Image:FormulaEa.jpg]] |
| − | raw termination efficiency, that not consider the mCherry contribution | + | raw termination efficiency, that does not consider the mCherry contribution [[Image:FormulaEr.jpg]] |
| + | |||
| + | relative increase in the upstream gene expression, [[Image:RIequation.png]] | ||
| − | |||
where | where | ||
| Line 76: | Line 57: | ||
-''Vs'' is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker | -''Vs'' is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker | ||
| − | -''Vc'' is the A206K Venus peak’s intensity of the control construct | + | -''Vc'' is the A206K Venus peak’s intensity of the control construct without intervening terminator |
-''Cs'' is the mCherry peak’s intensity of the construct with the terminator inserted | -''Cs'' is the mCherry peak’s intensity of the construct with the terminator inserted | ||
| Line 83: | Line 64: | ||
| − | + | '''Measurements''' | |
| − | <div style="text-align:center">[[Image: | + | Our experiments exploited an ''E. coli'' lysogen strain carrying T7 RNA polymerase and lacIq. Additionally, the cells, i.e. ''E. coli'' BL21(DE3) pLysS, also contained a plasmid encoding T7 lysozyme and chloramphenicol resistance. T7 lysozyme is a natural inhibitor of T7 RNA polymerase activity, thus reducing background expression of the target genes. The T7 RNA polymerase is behind a lacUV5 promoter. |
| − | < | + | |
| − | text-align: | + | |
| + | |||
| + | <div style="text-align:center">[[Image:tacpromoterinvivo.jpg]]</div> | ||
| + | <div style="text-align:center">[[Image:Ptactabinvivo.jpg]]</div> | ||
| − | |||
| − | |||
<p style="width:600px; margin-left:150px; margin-bottom:60px; | <p style="width:600px; margin-left:150px; margin-bottom:60px; | ||
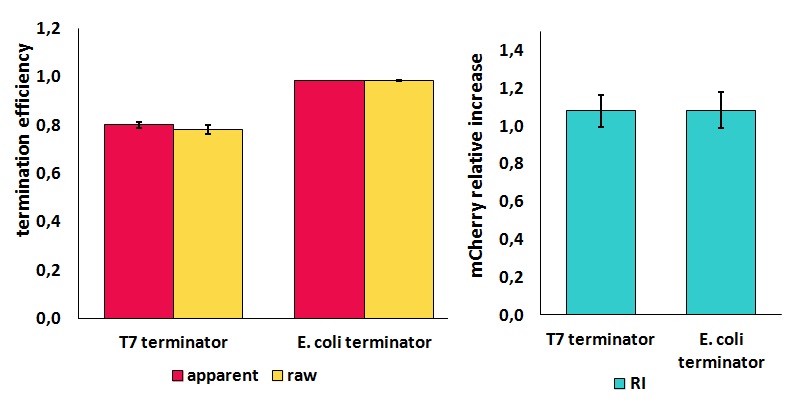
| − | text-align:justify "><em><strong>FIGURE | + | text-align:justify "><em><strong>FIGURE 2.</strong> '''Results obtained from the ''in vivo'' measurements of [[Part:BBa_K731721|BBa_K731721]] (T7 terminator) and [[Part:BBa_K731722|BBa_K731722]] (''E. coli'' terminator) using BBa_K731710'''<br/>The data here analyzed were collected from cells that were expressed for 4 h with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). |
| + | To identify optimal conditions, we screened samples that were and were not sonicated, sample dilutions to decrease the scattering of light, IPTG concentration, time of induction and time after induction. We found that the best results were obtained with induction with 0.5 mM IPTG for 3 h, followed by sonication and centrifugation. The cleared supernatant was then diluted 2-fold with phosphate buffer saline (PBS, 0.0067M PO4 without Ca or Mg, Cat number BE17-516F) and stored overnight at 4 °C. Finally, fluorescence was measured with a Varian Cary Eclipse spectrofluorimeter using emission and excitation wavelengths described above, a slit of 5nm and a voltage of 520V. The samples were left overnight at 4 °C to allow for sufficient time for the fluorescent proteins to properly mature (i.e. protein folding and chromophore formation). Each measurement was made in quadruplicate from different colonies from different transformations and from different plates. | ||
</em> </p> | </em> </p> | ||
| + | It is worth to highlight that the apparent and the raw transcriptional termination efficiencies, shown in figure 2, are essentially different from both a mathematical and a biological point of view. More specifically, raw termination efficiency is determined just by the expression of the downstream gene, while the apparent termination efficiency represents the variations that occur in the expression of both genes up- and down-stream of the terminator. They are in the same chart just for a comparison purpose. | ||
| + | |||
| + | For more information about the protocol we adopted to take these ''in vivo'' measurements, see [[BBa K731700 and BBa K731710 measurements]]. | ||
| + | |||
| + | |||
| + | |||
| + | We submitted also this backbone with each terminator used in this study inserted in the cloning region as measurement plasmids; we hope that this could be helpful for anyone who want to verify and further delve into our results. They are [[Part:BBa_K731711|BBa_K731711]] for the T7 terminator and [[Part:BBa_K731712|BBa_K731712]] for the ''E. coli'' terminator. | ||
| − | + | More information about the procedure used, the results obtained and our considerations can be found in the [http://2012.igem.org/Team:UNITN-Trento| Trento iGEM 2012 wiki page]. | |
| − | + | ==References== | |
| + | <biblio> | ||
| + | #Nojima pmid=16379390 | ||
| + | #Abe pmid=9150882 | ||
| + | </biblio> | ||
Latest revision as of 16:29, 11 October 2012
Platform for terminators analysis under the control of tac promoter
This is a platform for the characterization of terminator efficiency by fluorimetric measurements and ratiometric analysis under IPTG inducible tac promoter’s control. It was built starting from BBa_K731700, where the T7 promoter was replaced with the tac promoter by insertion/deletion mutagenesis. The platform has been used for the analysis of T7 wild type terminator's (BBa_K731721) and an E. coli terminator's (BBa_K731722) effects on protein synthesis.
The combined use of BBa_K731710 and BBa_K731700 allows to analyze also any potential difference in terminators' activity due to different RNA polymerases.
Usage and Biology
We made some preliminary analyses to define the best procedure to use this platform. We found that the emission from mVenus was much stronger than the mCherry emission, resulting in some spectral overlap. To improve the quality of the data, off-peak excitation and emission wavelengths were identified that minimized the effect. Therefore, mVenus was excitated at 485 nm (excitation peak is at 515 nm), and the emission of mVenus was collected at its maximum position, i.e. 528 nm. The maximum mCherry excitation wavelength was used (587 nm), but the emission of mCherry was read at 615 nm, as opposed to the maximum at 609 nm. Here is a summary of these wavelengths:
| Standard Excitation (nm) | Standard Emission (nm) | Modified Excitation (nm) | Modified Emission (nm) | |
| mCherry | 587 | 609 | 587 | 615 |
| mVenus | 515 | 528 | 485 | 528 |
TABLE 1. Standard and modified excitation and emission wavelengths.
This part was used to characterize a T7 wild type terminator (BBa_K731721) and an E. coli terminator (BBa_K731722).
These terminators's activity was measured as the ratio between the two proteins' levels using the equation found in literature for termination efficiency.
Doing this measurements, however, we realized that terminators can have an effect also on mCherry expression, enhancing it. This effect was previously described Abe. As a consequence of this unexpected outcome, we defined other two parameters that are here summarized in addition to the literature definition of termination efficiency.
The parameters used to analyze the data are:
apparent termination efficiency, calculated with the equation found in literature Nojima, 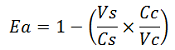
raw termination efficiency, that does not consider the mCherry contribution 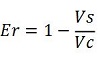
relative increase in the upstream gene expression, 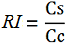
where
-Vs is the A206K Venus peak’s intensity of the construct with the terminator of interest inserted in the prefix-suffix linker
-Vc is the A206K Venus peak’s intensity of the control construct without intervening terminator
-Cs is the mCherry peak’s intensity of the construct with the terminator inserted
-Cc is the mCherry peak’s intensity of the control construct
Measurements
Our experiments exploited an E. coli lysogen strain carrying T7 RNA polymerase and lacIq. Additionally, the cells, i.e. E. coli BL21(DE3) pLysS, also contained a plasmid encoding T7 lysozyme and chloramphenicol resistance. T7 lysozyme is a natural inhibitor of T7 RNA polymerase activity, thus reducing background expression of the target genes. The T7 RNA polymerase is behind a lacUV5 promoter.
FIGURE 2. Results obtained from the in vivo measurements of BBa_K731721 (T7 terminator) and BBa_K731722 (E. coli terminator) using BBa_K731710
The data here analyzed were collected from cells that were expressed for 4 h with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG).
To identify optimal conditions, we screened samples that were and were not sonicated, sample dilutions to decrease the scattering of light, IPTG concentration, time of induction and time after induction. We found that the best results were obtained with induction with 0.5 mM IPTG for 3 h, followed by sonication and centrifugation. The cleared supernatant was then diluted 2-fold with phosphate buffer saline (PBS, 0.0067M PO4 without Ca or Mg, Cat number BE17-516F) and stored overnight at 4 °C. Finally, fluorescence was measured with a Varian Cary Eclipse spectrofluorimeter using emission and excitation wavelengths described above, a slit of 5nm and a voltage of 520V. The samples were left overnight at 4 °C to allow for sufficient time for the fluorescent proteins to properly mature (i.e. protein folding and chromophore formation). Each measurement was made in quadruplicate from different colonies from different transformations and from different plates.
It is worth to highlight that the apparent and the raw transcriptional termination efficiencies, shown in figure 2, are essentially different from both a mathematical and a biological point of view. More specifically, raw termination efficiency is determined just by the expression of the downstream gene, while the apparent termination efficiency represents the variations that occur in the expression of both genes up- and down-stream of the terminator. They are in the same chart just for a comparison purpose.
For more information about the protocol we adopted to take these in vivo measurements, see BBa K731700 and BBa K731710 measurements.
We submitted also this backbone with each terminator used in this study inserted in the cloning region as measurement plasmids; we hope that this could be helpful for anyone who want to verify and further delve into our results. They are BBa_K731711 for the T7 terminator and BBa_K731712 for the E. coli terminator.
More information about the procedure used, the results obtained and our considerations can be found in the [http://2012.igem.org/Team:UNITN-Trento| Trento iGEM 2012 wiki page].
References
<biblio>
- Nojima pmid=16379390
- Abe pmid=9150882
</biblio>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 6865
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 6871 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 6865
Illegal BglII site found at 6011
Illegal BamHI site found at 6847
Illegal XhoI site found at 753 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 6865
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 6865
Plasmid lacks a suffix.
Illegal XbaI site found at 6880
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 714
Illegal NgoMIV site found at 1051
Illegal NgoMIV site found at 4231
Illegal NgoMIV site found at 4391
Illegal NgoMIV site found at 5979
Illegal AgeI site found at 6815 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI site found at 2228
Illegal SapI.rc site found at 3310