Difference between revisions of "Part:pSB2K3"
Smelissali (Talk | contribs) |
(→Usage and Biology) |
||
| (3 intermediate revisions by one other user not shown) | |||
| Line 4: | Line 4: | ||
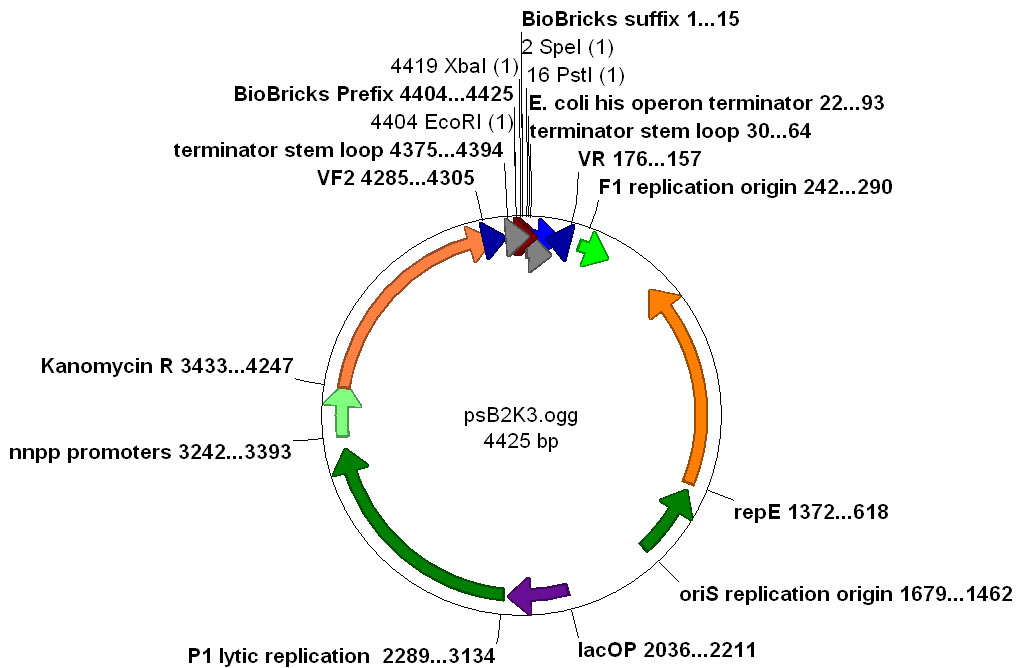
pSB2K3 is a variable copy number plasmid carrying kanamycin resistance. The plasmid contains the F' replication origin and also the [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 P1 lytic replication origin]. Genes encoding proteins that are required to activate the P1 lytic replication origin are carried on the plasmid under the control of a lacI-regulated promoter. When grown in cells producing LacI the plasmid is maintained at very low copy (<10 per cell). When induced with IPTG the P1 lytic replication proteins are produced and the P1 lytic replication origin takes over, raising the copy number to very high levels (>100++ per cell). The copy control features of pSB2K3 are useful for cloning "difficult" parts while still being able to prepare large amounts of DNA. | pSB2K3 is a variable copy number plasmid carrying kanamycin resistance. The plasmid contains the F' replication origin and also the [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 P1 lytic replication origin]. Genes encoding proteins that are required to activate the P1 lytic replication origin are carried on the plasmid under the control of a lacI-regulated promoter. When grown in cells producing LacI the plasmid is maintained at very low copy (<10 per cell). When induced with IPTG the P1 lytic replication proteins are produced and the P1 lytic replication origin takes over, raising the copy number to very high levels (>100++ per cell). The copy control features of pSB2K3 are useful for cloning "difficult" parts while still being able to prepare large amounts of DNA. | ||
| − | Additionally, pSB2K3 has terminators bracketing its MCS which are designed to prevent transcription from *inside* the MCS from reading out into the vector. The efficiency of these terminators is known to be < 100%. Ideally we would construct a future set of terminators for bracketing a MCS that were 100% efficient in terminating both into and out of the MCS region. | + | Additionally, pSB2K3 has terminators bracketing its MCS ("multi-cloning site") which are designed to prevent transcription from *inside* the MCS from reading out into the vector. The efficiency of these terminators is known to be < 100%. Ideally we would construct a future set of terminators for bracketing a MCS that were 100% efficient in terminating both into and out of the MCS region. |
| + | Annotated [http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] file for [https://static.igem.org/mediawiki/parts/2/22/PsB2K3.ogg pSB2K3] [[Image:PSB2K3 map.jpg|thumb|center|Plasmid map for pSB2K3]] | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 11: | Line 12: | ||
*** Copy number control & induction via IPTG *** | *** Copy number control & induction via IPTG *** | ||
| − | Cells that contain a chromosomal lacIq allelle (e.g., [[Part:BBa_V1003|E.coli D1210]], an HB101 derivative) provide tight repression of the P1 replication origin (NOTE: since pSCANS uses an F' origin the lacIq allele can't be carried on a separate F-based plasmid). Induction to high copy can be carried out by addition of IPTG (1mM final concentration). IPTG can be added at different points during growth (to suit your application). For instance, to prepare both cells (e.g., for glycerol stocks) and DNA (e.g., for further cloning) grow the cells to mid log, pull a sample for the glycerol stock, add IPTG and continue growth, prepare DNA from a saturated culture. | + | Cells that contain a chromosomal lacIq allelle (e.g., [[Part:BBa_V1003|E.coli D1210]], an HB101 derivative) provide tight repression of the P1 replication origin (NOTE: since pSCANS uses an F' origin the lacIq allele can't be carried on a separate F-based plasmid). Induction to high copy can be carried out by addition of IPTG (1mM final concentration). IPTG can be added at different points during growth (to suit your application). For instance, to prepare both cells (e.g., for glycerol stocks) and DNA (e.g., for further cloning) grow the cells to mid log, pull a sample for the glycerol stock, add IPTG and continue growth, prepare DNA from a saturated culture. |
| + | |||
| + | Result from experiments shows that for IPTG concentration less than 20 μmol/L, the effect of inducement is scarcely perceivable (mean copy number: 1~3). It also shows that from 20 μmol/L to 100 μmol/L the copy number is induced by IPTG in a way quite proportional to its concentration. It seems that over 100 μmol/L IPTG concentration is “saturated” and a further increase in IPTG concentration cannot raise the copy number (mean copy number: 150±10 in exponentially growing culture). | ||
| + | |||
| + | This result is from exponentially growing culture. For cell culture in stationary phase the mean copy number possibly increases. | ||
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 repE] gene product controls the replication initiation of the plasmid via the F1 replication origin. | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 repE] gene product controls the replication initiation of the plasmid via the F1 replication origin. | ||
| + | |||
| + | Primers for these Biobrick vectors can be found in [[Part:BBa_G00100]] (aka VF2) and [[Part:BBa_G00101]] (aka. VR) | ||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 13:43, 4 November 2008
Inducible copy number BioBrick plasmid
pSB2K3 is a variable copy number plasmid carrying kanamycin resistance. The plasmid contains the F' replication origin and also the [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 P1 lytic replication origin]. Genes encoding proteins that are required to activate the P1 lytic replication origin are carried on the plasmid under the control of a lacI-regulated promoter. When grown in cells producing LacI the plasmid is maintained at very low copy (<10 per cell). When induced with IPTG the P1 lytic replication proteins are produced and the P1 lytic replication origin takes over, raising the copy number to very high levels (>100++ per cell). The copy control features of pSB2K3 are useful for cloning "difficult" parts while still being able to prepare large amounts of DNA.
Additionally, pSB2K3 has terminators bracketing its MCS ("multi-cloning site") which are designed to prevent transcription from *inside* the MCS from reading out into the vector. The efficiency of these terminators is known to be < 100%. Ideally we would construct a future set of terminators for bracketing a MCS that were 100% efficient in terminating both into and out of the MCS region.
Annotated [http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] file for pSB2K3Usage and Biology
- Copy number control & induction via IPTG ***
Cells that contain a chromosomal lacIq allelle (e.g., E.coli D1210, an HB101 derivative) provide tight repression of the P1 replication origin (NOTE: since pSCANS uses an F' origin the lacIq allele can't be carried on a separate F-based plasmid). Induction to high copy can be carried out by addition of IPTG (1mM final concentration). IPTG can be added at different points during growth (to suit your application). For instance, to prepare both cells (e.g., for glycerol stocks) and DNA (e.g., for further cloning) grow the cells to mid log, pull a sample for the glycerol stock, add IPTG and continue growth, prepare DNA from a saturated culture.
Result from experiments shows that for IPTG concentration less than 20 μmol/L, the effect of inducement is scarcely perceivable (mean copy number: 1~3). It also shows that from 20 μmol/L to 100 μmol/L the copy number is induced by IPTG in a way quite proportional to its concentration. It seems that over 100 μmol/L IPTG concentration is “saturated” and a further increase in IPTG concentration cannot raise the copy number (mean copy number: 150±10 in exponentially growing culture).
This result is from exponentially growing culture. For cell culture in stationary phase the mean copy number possibly increases.
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=YP_006518.1 repE] gene product controls the replication initiation of the plasmid via the F1 replication origin.
Primers for these Biobrick vectors can be found in Part:BBa_G00100 (aka VF2) and Part:BBa_G00101 (aka. VR)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4404
Illegal NheI site found at 3317
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 4410 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4404
Illegal BglII site found at 1618
Illegal BglII site found at 2665
Illegal XhoI site found at 3462 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 4404
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 4404
Plasmid lacks a suffix.
Illegal XbaI site found at 4419
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 1061
Illegal NgoMIV site found at 3303
Illegal AgeI site found at 937
Illegal AgeI site found at 2596 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.