Difference between revisions of "Part:BBa B0032"
(→Functional Parameters: Austin_UTexas) |
|||
| (7 intermediate revisions by 5 users not shown) | |||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
== IIT Madras 2016's Characterization == | == IIT Madras 2016's Characterization == | ||
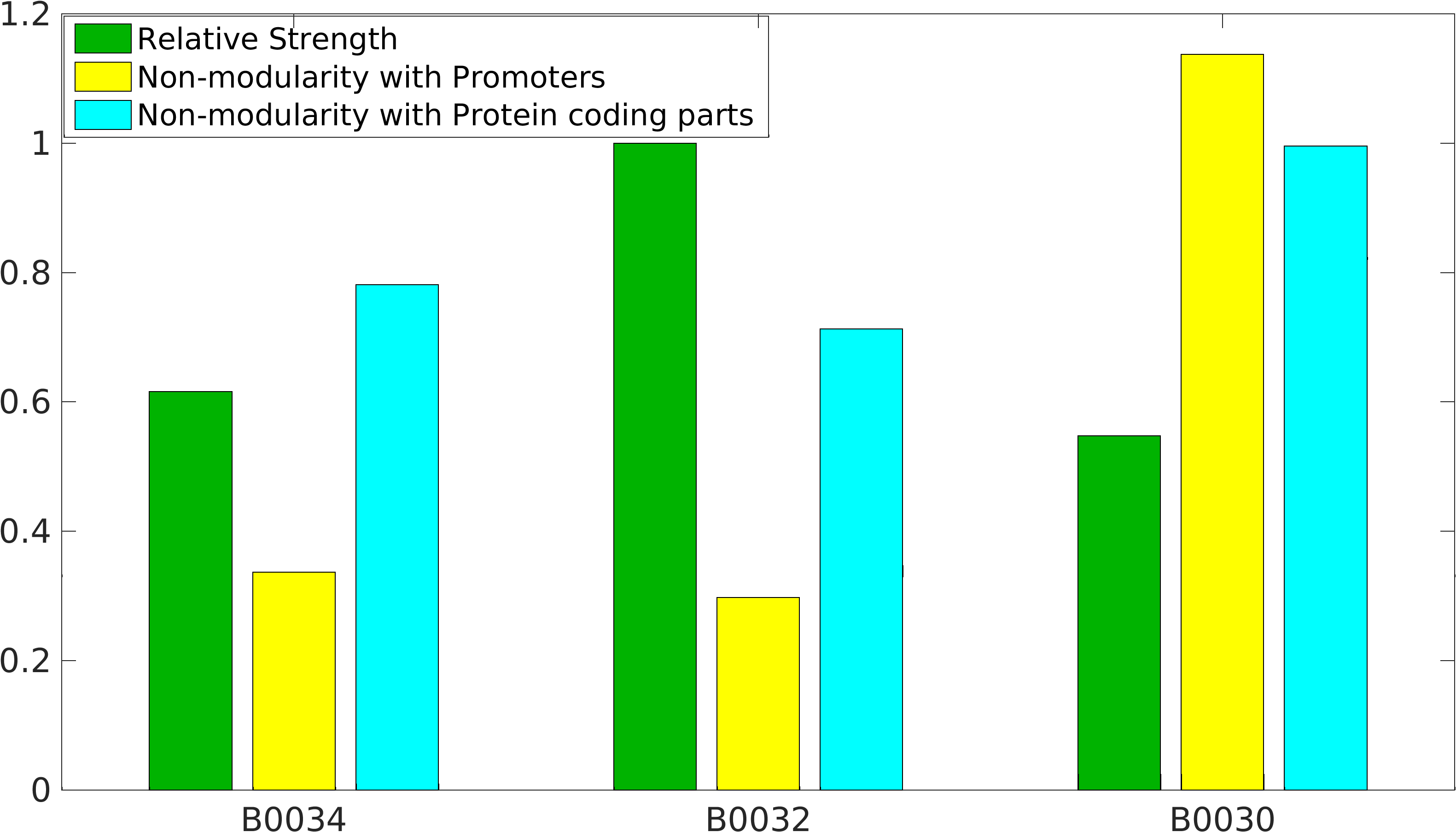
[[Image:Nonmodularity_of_B343230.png|400px|right|Characterization of popular BioBrick RBSs]] | [[Image:Nonmodularity_of_B343230.png|400px|right|Characterization of popular BioBrick RBSs]] | ||
| Line 12: | Line 13: | ||
<p>Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]</p> | <p>Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]</p> | ||
== Experimentation == | == Experimentation == | ||
| − | <p>Biobrick RBSs [https://parts.igem.org/Part:BBa_K1956022 B0030], [https://parts.igem.org/Part:BBa_K1956023 B0031], [https://parts.igem.org/Part:BBa_K1956014 B0032], [https://parts.igem.org/Part:BBa_K1956015 B0034] were used in our 'Noise in Device' experiment to understand the role of RBS parts in noise.</p> | + | <p>Biobrick RBSs [https://parts.igem.org/Part:BBa_K1956022 B0030], [https://parts.igem.org/Part:BBa_K1956023 B0031], [https://parts.igem.org/Part:BBa_K1956014 B0032], [https://parts.igem.org/Part:BBa_K1956015 B0034] were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.</p> |
<!-- --> | <!-- --> | ||
| Line 24: | Line 25: | ||
<html><hr><h3 style="color:ltblue;"><a href="http://2010.igem.org/Team:Warsaw/Stage1/RBSMeas">Team Warsaw 2010's measurement</a></h3> | <html><hr><h3 style="color:ltblue;"><a href="http://2010.igem.org/Team:Warsaw/Stage1/RBSMeas">Team Warsaw 2010's measurement</a></h3> | ||
RBS strength (relative to <a href="https://parts.igem.org/Part:BBa_B0034">B0034</a>): 33,96% | RBS strength (relative to <a href="https://parts.igem.org/Part:BBa_B0034">B0034</a>): 33,96% | ||
| + | </html> | ||
| + | ===Contribution=== | ||
| + | Group: Valencia_UPV iGEM 2018 | ||
| + | <br> | ||
| + | Author: Adrián Requena Gutiérrez, Carolina Ropero | ||
| + | <br> | ||
| + | Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS. | ||
| + | <br> | ||
| + | Documentation: | ||
| + | <br> | ||
| + | BBa_K2656010 is the [https://parts.igem.org/Part:BBa_B0032 BBa_B0032] ribosome binding site standardized into the Golden Gate assembly method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure the correct spacing for the CDS expression when assembled into a TU. | ||
| + | |||
| + | Characterization of this part was performed with the transcriptional unit [https://parts.igem.org/Part:BBa_K2656102 BBa_K2656102], which was used in a comparative RBS expression experiment with composite parts [https://parts.igem.org/Part:BBa_K2656103 BBa_K2656103] and [https://parts.igem.org/Part:BBa_K2656101 BBa_K2656101]. | ||
| + | They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, coding sequence and terminator. | ||
| + | |||
| + | By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS tested. | ||
| + | |||
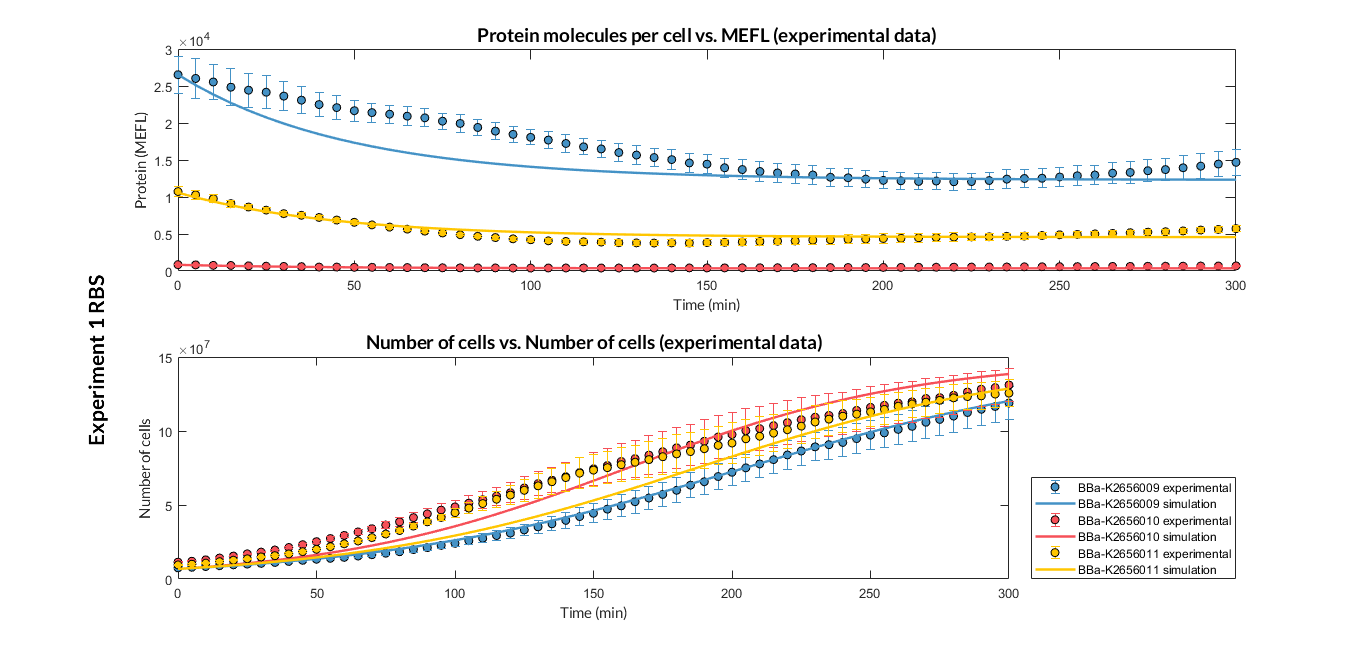
| + | [[File:T--Valencia_UPV--optimization_exp1_RBS_graphUPV2018.png|900px|thumb|none|alt=RBS experiment 1.|Figure 1. RBS expression experiment with K2656009, K26560010 and K2656011 RBS basic parts]] | ||
| + | |||
| + | {|class='wikitable' | ||
| + | |colspan=4|Table 1. Optimized parameters for the BBa_K2656010 RBS. | ||
| + | |- | ||
| + | |'''Parameter''' | ||
| + | |'''Value''' | ||
| + | |- | ||
| + | |Translation rate p | ||
| + | |p = 0.01 min-1 | ||
| + | |- | ||
| + | |Dilution rate μ | ||
| + | | μ = 0.01641 min-1 | ||
| + | |} | ||
| + | |||
| + | We have also calculated the relative force between the different RBS, taking [https://parts.igem.org/Part:BBa_K2656009 BBa_K2656009 strong RBS] as a reference. It has been defined as the quotient between the values of the protein in equilibrium of the results of the simulation of one RBS and another reference RBS. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated. | ||
| + | |||
| + | {|class='wikitable' | ||
| + | |colspan=4|Table 2. BBa_K2656010 (GB B0030 RBS) relative strength and p ratio. | ||
| + | |- | ||
| + | |'''Parameter''' | ||
| + | |'''Value''' | ||
| + | |- | ||
| + | |Relative strength | ||
| + | |0.045 | ||
| + | |- | ||
| + | |p parameter ratio (pRBS/pref) | ||
| + | |0.048 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | >Internal Priming Screening Characterization of BBa_B0032: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer. | ||
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_B0032 parameters</partinfo> | ||
| + | (Relative to <partinfo>B0034</partinfo>) | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 3.0 ± 6.2% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
</html> | </html> | ||
Latest revision as of 19:10, 3 September 2020
RBS.3 (medium) -- derivative of BBa_0030
Weak1 RBS based on Ron Weiss thesis. Strength is considered relative to BBa_B0030, BBa_B0031, BBa_B0033.
Usage and Biology
IIT Madras 2016's Characterization
Modelling
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]
Experimentation
Biobrick RBSs B0030, B0031, B0032, B0034 were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| biology | -NA- |
| efficiency | 0.3 |
(Relative to BBa_B0034)
Team Warsaw 2010's measurement
RBS strength (relative to B0034): 33,96%Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS.
Documentation:
BBa_K2656010 is the BBa_B0032 ribosome binding site standardized into the Golden Gate assembly method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure the correct spacing for the CDS expression when assembled into a TU.
Characterization of this part was performed with the transcriptional unit BBa_K2656102, which was used in a comparative RBS expression experiment with composite parts BBa_K2656103 and BBa_K2656101. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, coding sequence and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS tested.
| Table 1. Optimized parameters for the BBa_K2656010 RBS. | |||
| Parameter | Value | ||
| Translation rate p | p = 0.01 min-1 | ||
| Dilution rate μ | μ = 0.01641 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. It has been defined as the quotient between the values of the protein in equilibrium of the results of the simulation of one RBS and another reference RBS. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656010 (GB B0030 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.045 | ||
| p parameter ratio (pRBS/pref) | 0.048 | ||
>Internal Priming Screening Characterization of BBa_B0032: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.