Difference between revisions of "Part:BBa C0060"
| (40 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_C0060 short</partinfo> | <partinfo>BBa_C0060 short</partinfo> | ||
| − | Coding region for the autoinducer inactivation enzyme A (<em>aiiA</em>) LVA | + | Coding region for the autoinducer inactivation enzyme A (<em>aiiA</em>) [[Help:Tag|LVA tagged]] (for untagged version see [[Part:BBa_C0160]]. The gene was originally isolated from <em>Bacillus</em> sp. 240B1 and it encodes an enzyme that catalyzes the degradation of N-acyl-homoserine lactones ([[Acyl-HSLs|AHLs]])--quorum sensing autoinducers. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | The | + | The <em>aiiA</em> gene has 750 nucleotides and encodes a protein of 250aa with a predicted molecular mass of 28,036Da and an isoelectric point of 4.7. The protein has no hydrophobic signal peptide at the <em>N</em>-terminus and therefore it is believed that it is not secreted. This is supported by the observation that when <em>aiiA </em> is expressed in <em>E.coli</em> [[Part:BBa_V1001|DH5alpha]] or <em>Bacillus</em> 240B1 cells no autoinducer inactivation is detected in the supernatants of these cultures. The AiiA protein has no significant overall homology with other known proteins, but based on the presence of two well-conserved motifs it is believed that it is a metalloenzyme. |
| + | |||
| + | ===Characterization of BBa_C0060 by Tec-Chihuahua 2017=== | ||
| + | |||
| + | <p align= "justify">One of the fundamental Biobricks® of this project is the aiiA gene, it codifies for an enzyme that catalyzes the degradation of N-Acyl homoserine lactones, which are quorum sensing autoinducers. The part was published on iGEM's Parts Registry in 2013, but up until now, it's not known whether this gene can be expressed in <i>Erwinia amylovora</i> or not. Two types of aiiA were used in this project: the one published in iGEM's catalog, and the one that we synthesized thanks to sponsorship granted by IDT®. The sequence of the first mentioned is reported as being putative, while the latter's confirmed to codify for our protein of interest; nonetheless, no information is known about the expression of either in <i>Erwinia amylovora</i>. The contribution of this characterization to the existing part (BBa_C0060) as a bronze requisite is supported by the work done with it within the project. In the end, it was concluded that our composite BioBrick® https://parts.igem.org/Part:BBa_K2471000 (BBa_K2471000) that was created in order to express the Registry's aiiA, successfully works in <i>Erwinia amylovora</i>.</p> | ||
| + | |||
| + | <p align= "justify">The characterization of BBa_C0060 was carried out, first, by creating a composite part https://parts.igem.org/Part:BBa_K2471000 (BBa_K2471000) capable of expressing the Biobrick® of interest; this was done by the addition of a T7 promoter and RBS https://parts.igem.org/Part:BBa_K525998 (BBa_K525998) and a T1 terminator https://parts.igem.org/Part:BBa_B0010 (BBa_B0010) to the basic part. The design was first done in silico utilizing SnapGene®, where the program indicated that the plasmid would have a length of 3,010 base pairs. After verifying the feasibility of creating this BioBrick®, the part was synthesized using the 3A assembly method. Agarose gel (1%) electrophoresis was performed to corroborate that the obtained molecular weight coincided with the expected one.</p><br><br> | ||
| + | |||
| + | <center>[[File:Snapgeneaiia.png|500px]]</center> | ||
| + | <center><div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 1. </b>(A) SnapGene® map of BBa_K2471000. (B) Agarose gel (1%) electrophoresis of BBa_K2471000 compared with NEB Quick-Load® Purple 2-Log DNA Ladder (0.1 - 10.0 kb), where the highlighted bands correspond to approximately 3,000 base pairs.</sub></center> | ||
| + | </div></div> | ||
| + | |||
| + | <p align= "justify">The bacteria <i>Erwinia amylovora</i> was transformed by electroporation, with this new BioBrick®. Electrophoresis in a polyacrylamide gel (12%) was performed in order to corroborate that the protein of interest was indeed expressed, as this is what was theoretically proposed based on the previous biographical review. The general methodology (to see specifications visit protocols section) involved a growth curve of the bacteria; also, a sample was taken every hour (0 - 8 hours) and stored in refrigeration to later perform total protein extraction; the protein molecular weight marker used was BIO-RAD Precision Plus Protein™ Dual Color Standards. | ||
| + | |||
| + | To calculate the molecular weight of the protein, the sequence provided by the iGEM Parts Registry for BBa_C0060 was taken and introduced in ExPASy - Translate tool, which generated the open reading frame in amino acids. This peptidic sequence was then analyzed in ExPASy - ProtParam tool, where a molecular weight of 30.32026 kDa was calculated.</p> | ||
| + | |||
| + | [[File:Sdsaiia.png|500px]] | ||
| + | <div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 2. </b> SDS-PAGE (12%) of <i>Erwinia amylovora</i> transformed with BBa_K2471000.</sub> | ||
| + | </div></div><br> | ||
| + | |||
| + | <p align= "justify">As it can be seen in Figure 1, there is an appreciable band present at the approximate weight of 30 kDa from the fourth hour onwards. As a point of comparison, SDS-PAGE of wild <i>Erwinia amylovora</i> was also carried out. However, no banding was seen whatsoever. We think that this happened because the protocol used is not specifically designed for this bacteria, and since <i>Erwinia amylovora</i> forms a very complex biofilm (Koczan et al., 2009), the cell lysis was not carried out correctly; and thus, neither did the protein extraction. In like manner, we propose that the SDS-PAGE shown in figure 1, where the protein of interest is expressed, was done correctly because of the protein's function, which is quorum quenching; this affects the production of important components of its biofilm, which consequently leads to its deficient production. When reviewing the literature on this, not much was found, <i>Erwinia amylovora</i> is a poorly studied bacteria. </p> | ||
| + | |||
| + | <p align= "justify">Nonetheless, a comparison was made with an SDS-PAGE that was executed using the same methodology, but utilizing <i>Erwinia amylovora</i> transformed with BBa_K2471003 instead. As BBa_C0060 is registered as being a putative N-Acyl homoserine lactonase aiiA, this new BioBrick contains the genetic circuitry to correctly express our protein, whose sequence (that has small changes compared to BBa_C0060, with a molecular weight of 29.45340 kDa) is confirmed to indeed codify for a N-Acyl homoserine lactonase aiiA. This part is a new addition to the Parts Registry, more information about it can be found in the silver section of our results.</p><br><br> | ||
| + | |||
| + | [[File:Sdsaiia2.png|500px]] | ||
| + | <div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 3. </b> SDS-PAGE (12%) of <i>Erwinia amylovora</i> transformed with BBa_K2471003.</sub> | ||
| + | </div></div><br> | ||
| + | </center> | ||
| + | <p align= "justify">In the same way as Figure 1, Figure 3 shows an appreciable band present at the approximate weight of 30 kDa, starting from the fourth hour onwards; meaning that this band is indeed the enzyme aiiA. With this, we establish that we characterized BBa_C0060 by proving that <i>Erwinia amylovora</i> can indeed express this protein.</p> | ||
| + | |||
| + | <h4>Inoculation of Apple Trees</h4> | ||
| + | |||
| + | <p align= "justify"> Two different types of <i>Erwinia amylovora</i>, wild and transformed with our composite aiiA BioBrick (BBa_K2471000), were inoculated on apple leaves. The disease (Fire Blight) began to be visible three days after inoculation and developed in a gradual and similar manner until the eighth day when the disease considerably increased its expansion through the leaf (Figure 1). However, differences in disease development (both the severity and onset time) were seen between the wild-type <i>E. amylovora </i>(Figure 2) and the one transformed with the aiiA BioBrick (Figure 3). The assay consisted of cutting leaves and inoculating them with the previously mentioned types of <i>E. amylovora</i> -plus a negative control, which was inoculated with medium without inoculum-, in order to simulate close to real conditions and see how each of them evolved.</p><br><br><br> | ||
| + | <center> | ||
| + | [[File:Hojas1.png|500px]] | ||
| + | <div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 4. </b> Evolution of leaf decay in the negative control.</sub> | ||
| + | </div></div><br> | ||
| + | |||
| + | [[File:Hojasyhjh.png|500px]] | ||
| + | <div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 5. </b> Evolution of leaf necrosis through time caused by the inoculation of a wild-type E. amylovora.</sub> | ||
| + | </div></div><br> | ||
| + | |||
| + | |||
| + | [[File:Hojas3.png|500px]] | ||
| + | <div class="row"> | ||
| + | <div class="col-md-8"> | ||
| + | <sub><b>Figure 6. </b> Evolution of leaf deterioration through time in a leaf inoculated with aiiA E. amylovora. Phenotypic dismissal of leaf necrosis is visualized, in comparison with Figure 2. </sub> | ||
| + | </div></div><br> | ||
| + | </center> | ||
| + | <p align= "justify">Erwinia amylovora’s virulence factors principally depend on its cellular density, N-Acyl homoserine lactones (AHLs) are responsible for and involved in the synthesis of many secondary metabolites, among them, exopolysaccharides like the amylovoran and levan. AHLs in E. amylovora has been proven to contribute to the expression of several virulence factors and disease development; similar to the role they play in other pathogenic bacteria. When incorporating the aiiA BioBrick (BBa_K2471000) into E. amylovora, leaf necrosis was inhibited (Figure 3), when compared with the wild-type E. amylovora (Figure 2). The production of the characteristic bacterial exudate of the disease was inhibited, which is composed of the amylovoran and levan EPS, and the infection that is presented by the type III secretion system dismissed. </p> | ||
| + | |||
| + | <p align= "justify">The gene expressed is an N-Acyl homoserine lactonase, which is involved in the hydrolysis of AHLs. The degradation of these molecules stops the AHL-EamR complex from binding, which in turn, stops the activation of an operon responsible for the EamI transcription of intracellular AHLs and other genes that contribute to several virulence factors. </p> | ||
| + | |||
| + | <p align= "justify">Phenotypic dismissal of Fire Blight was observed when the disease was caused by E. amylovora electroporated with our aiiA BioBrick compared to the wild-type E. amylovora. These results show a noticeable decrease in disease development in the bacteria transformed with our BioBrick (Figure 3). Thus, we can conclude that our gene does reduce the decay caused by the disease in close-to-reality scenarios.</p><br><br><br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 14: | Line 76: | ||
| − | + | ||
| − | + | >Internal Priming Screening Characterization of BBa_C0060: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer. | |
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | |||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
<partinfo>BBa_C0060 parameters</partinfo> | <partinfo>BBa_C0060 parameters</partinfo> | ||
| − | < | + | <h3><center>Burden Imposed by this Part:</center></h3> |
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: -0.4 ± 2.9% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 02:40, 25 August 2020
autoinducer inactivation enzyme from Bacillus; hydrolyzes acetyl homoserine lactone
Coding region for the autoinducer inactivation enzyme A (aiiA) LVA tagged (for untagged version see Part:BBa_C0160. The gene was originally isolated from Bacillus sp. 240B1 and it encodes an enzyme that catalyzes the degradation of N-acyl-homoserine lactones (AHLs)--quorum sensing autoinducers.
Usage and Biology
The aiiA gene has 750 nucleotides and encodes a protein of 250aa with a predicted molecular mass of 28,036Da and an isoelectric point of 4.7. The protein has no hydrophobic signal peptide at the N-terminus and therefore it is believed that it is not secreted. This is supported by the observation that when aiiA is expressed in E.coli DH5alpha or Bacillus 240B1 cells no autoinducer inactivation is detected in the supernatants of these cultures. The AiiA protein has no significant overall homology with other known proteins, but based on the presence of two well-conserved motifs it is believed that it is a metalloenzyme.
Characterization of BBa_C0060 by Tec-Chihuahua 2017
One of the fundamental Biobricks® of this project is the aiiA gene, it codifies for an enzyme that catalyzes the degradation of N-Acyl homoserine lactones, which are quorum sensing autoinducers. The part was published on iGEM's Parts Registry in 2013, but up until now, it's not known whether this gene can be expressed in Erwinia amylovora or not. Two types of aiiA were used in this project: the one published in iGEM's catalog, and the one that we synthesized thanks to sponsorship granted by IDT®. The sequence of the first mentioned is reported as being putative, while the latter's confirmed to codify for our protein of interest; nonetheless, no information is known about the expression of either in Erwinia amylovora. The contribution of this characterization to the existing part (BBa_C0060) as a bronze requisite is supported by the work done with it within the project. In the end, it was concluded that our composite BioBrick® https://parts.igem.org/Part:BBa_K2471000 (BBa_K2471000) that was created in order to express the Registry's aiiA, successfully works in Erwinia amylovora.
The characterization of BBa_C0060 was carried out, first, by creating a composite part https://parts.igem.org/Part:BBa_K2471000 (BBa_K2471000) capable of expressing the Biobrick® of interest; this was done by the addition of a T7 promoter and RBS https://parts.igem.org/Part:BBa_K525998 (BBa_K525998) and a T1 terminator https://parts.igem.org/Part:BBa_B0010 (BBa_B0010) to the basic part. The design was first done in silico utilizing SnapGene®, where the program indicated that the plasmid would have a length of 3,010 base pairs. After verifying the feasibility of creating this BioBrick®, the part was synthesized using the 3A assembly method. Agarose gel (1%) electrophoresis was performed to corroborate that the obtained molecular weight coincided with the expected one.
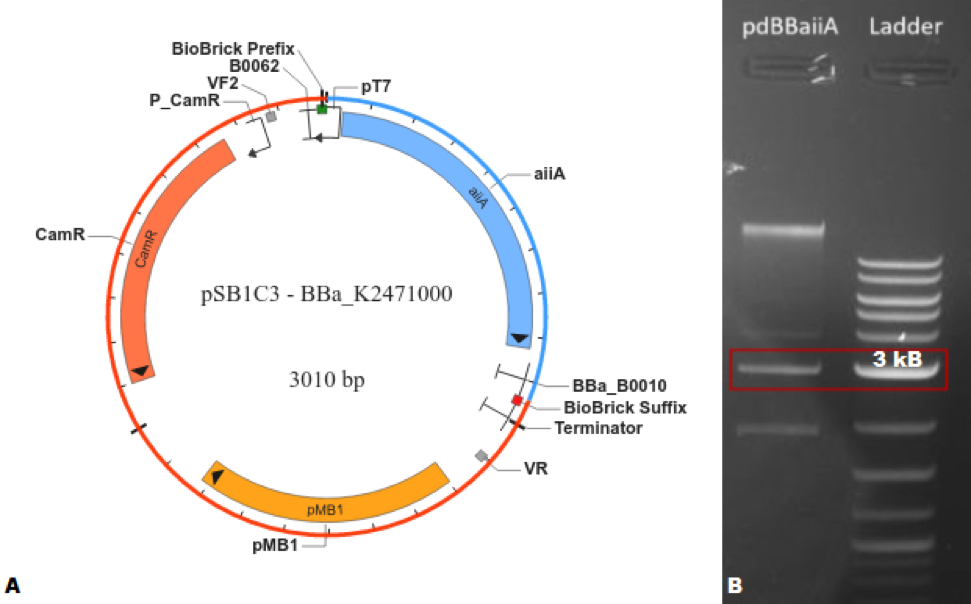
Figure 1. (A) SnapGene® map of BBa_K2471000. (B) Agarose gel (1%) electrophoresis of BBa_K2471000 compared with NEB Quick-Load® Purple 2-Log DNA Ladder (0.1 - 10.0 kb), where the highlighted bands correspond to approximately 3,000 base pairs.</center>
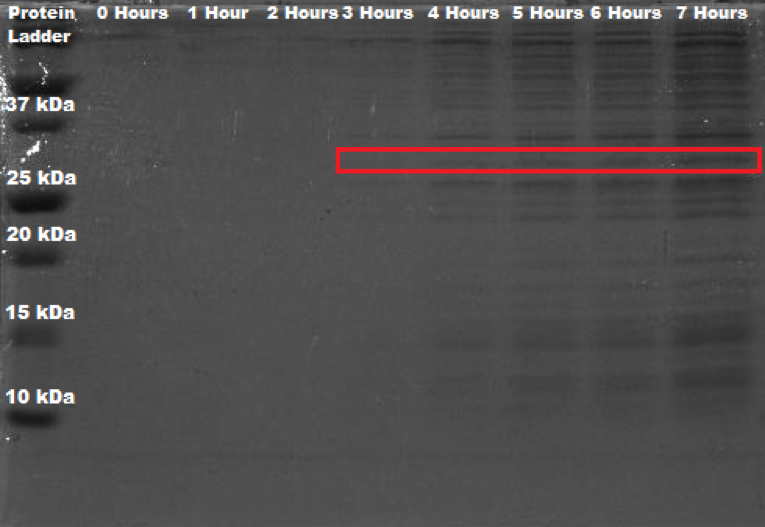
The bacteria Erwinia amylovora was transformed by electroporation, with this new BioBrick®. Electrophoresis in a polyacrylamide gel (12%) was performed in order to corroborate that the protein of interest was indeed expressed, as this is what was theoretically proposed based on the previous biographical review. The general methodology (to see specifications visit protocols section) involved a growth curve of the bacteria; also, a sample was taken every hour (0 - 8 hours) and stored in refrigeration to later perform total protein extraction; the protein molecular weight marker used was BIO-RAD Precision Plus Protein™ Dual Color Standards. To calculate the molecular weight of the protein, the sequence provided by the iGEM Parts Registry for BBa_C0060 was taken and introduced in ExPASy - Translate tool, which generated the open reading frame in amino acids. This peptidic sequence was then analyzed in ExPASy - ProtParam tool, where a molecular weight of 30.32026 kDa was calculated.
Figure 2. SDS-PAGE (12%) of Erwinia amylovora transformed with BBa_K2471000.
As it can be seen in Figure 1, there is an appreciable band present at the approximate weight of 30 kDa from the fourth hour onwards. As a point of comparison, SDS-PAGE of wild Erwinia amylovora was also carried out. However, no banding was seen whatsoever. We think that this happened because the protocol used is not specifically designed for this bacteria, and since Erwinia amylovora forms a very complex biofilm (Koczan et al., 2009), the cell lysis was not carried out correctly; and thus, neither did the protein extraction. In like manner, we propose that the SDS-PAGE shown in figure 1, where the protein of interest is expressed, was done correctly because of the protein's function, which is quorum quenching; this affects the production of important components of its biofilm, which consequently leads to its deficient production. When reviewing the literature on this, not much was found, Erwinia amylovora is a poorly studied bacteria.
Nonetheless, a comparison was made with an SDS-PAGE that was executed using the same methodology, but utilizing Erwinia amylovora transformed with BBa_K2471003 instead. As BBa_C0060 is registered as being a putative N-Acyl homoserine lactonase aiiA, this new BioBrick contains the genetic circuitry to correctly express our protein, whose sequence (that has small changes compared to BBa_C0060, with a molecular weight of 29.45340 kDa) is confirmed to indeed codify for a N-Acyl homoserine lactonase aiiA. This part is a new addition to the Parts Registry, more information about it can be found in the silver section of our results.
Figure 3. SDS-PAGE (12%) of Erwinia amylovora transformed with BBa_K2471003.
In the same way as Figure 1, Figure 3 shows an appreciable band present at the approximate weight of 30 kDa, starting from the fourth hour onwards; meaning that this band is indeed the enzyme aiiA. With this, we establish that we characterized BBa_C0060 by proving that Erwinia amylovora can indeed express this protein.
Inoculation of Apple Trees
Two different types of Erwinia amylovora, wild and transformed with our composite aiiA BioBrick (BBa_K2471000), were inoculated on apple leaves. The disease (Fire Blight) began to be visible three days after inoculation and developed in a gradual and similar manner until the eighth day when the disease considerably increased its expansion through the leaf (Figure 1). However, differences in disease development (both the severity and onset time) were seen between the wild-type E. amylovora (Figure 2) and the one transformed with the aiiA BioBrick (Figure 3). The assay consisted of cutting leaves and inoculating them with the previously mentioned types of E. amylovora -plus a negative control, which was inoculated with medium without inoculum-, in order to simulate close to real conditions and see how each of them evolved.
Figure 4. Evolution of leaf decay in the negative control.
Figure 5. Evolution of leaf necrosis through time caused by the inoculation of a wild-type E. amylovora.
Figure 6. Evolution of leaf deterioration through time in a leaf inoculated with aiiA E. amylovora. Phenotypic dismissal of leaf necrosis is visualized, in comparison with Figure 2.
Erwinia amylovora’s virulence factors principally depend on its cellular density, N-Acyl homoserine lactones (AHLs) are responsible for and involved in the synthesis of many secondary metabolites, among them, exopolysaccharides like the amylovoran and levan. AHLs in E. amylovora has been proven to contribute to the expression of several virulence factors and disease development; similar to the role they play in other pathogenic bacteria. When incorporating the aiiA BioBrick (BBa_K2471000) into E. amylovora, leaf necrosis was inhibited (Figure 3), when compared with the wild-type E. amylovora (Figure 2). The production of the characteristic bacterial exudate of the disease was inhibited, which is composed of the amylovoran and levan EPS, and the infection that is presented by the type III secretion system dismissed.
The gene expressed is an N-Acyl homoserine lactonase, which is involved in the hydrolysis of AHLs. The degradation of these molecules stops the AHL-EamR complex from binding, which in turn, stops the activation of an operon responsible for the EamI transcription of intracellular AHLs and other genes that contribute to several virulence factors.
Phenotypic dismissal of Fire Blight was observed when the disease was caused by E. amylovora electroporated with our aiiA BioBrick compared to the wild-type E. amylovora. These results show a noticeable decrease in disease development in the bacteria transformed with our BioBrick (Figure 3). Thus, we can conclude that our gene does reduce the decay caused by the disease in close-to-reality scenarios.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 99
- 1000COMPATIBLE WITH RFC[1000]
>Internal Priming Screening Characterization of BBa_C0060: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.