Difference between revisions of "Part:BBa K2926077"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2926077 short</partinfo> | <partinfo>BBa_K2926077 short</partinfo> | ||
| − | + | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 17: | ||
<partinfo>BBa_K2926077 parameters</partinfo> | <partinfo>BBa_K2926077 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | Chloramphenicol is an antibiotic which easily diffuses across cell membranes. | ||
| + | Once inside the cell it interferes in the protein biosynthesis pathway. | ||
| + | More precisely it binds to specific residues of the 23S rRNA of the 50S ribosomal subunit | ||
| + | and interrupts the formation of new peptide bonds. <br> | ||
| + | In 1947 chloramphenicol was first discovered in an isolate from <i>Streptomyces venezuelae</i>, | ||
| + | later that decade it became the first artificially produced antibiotic. (Pong 1979) | ||
| + | In every day life chloramphenicol serves as a back up broadband antibiotic which is used in case other | ||
| + | antibiotics can not be applied. Normally it is only used in case of need, since it comes with a broad range of side effects. | ||
| + | Nevertheless, chloramphenicol is often one of the ingredients in eye ointments for the treatment of conjunctivitis. (Edwards 2009) | ||
| + | Furthermore it is used to treat hazardous bacterial infections like plaque, cholera, MRSA or typhoid fever. (Ingebrigtsen 2017)<br> | ||
| + | Additionally, it is one of the most commonly used antibiotics in molecular biology for the selection of genetically modified organisms. | ||
| + | Many of the vectors for gene transfer used in research carry a gene encoding a chloramphenicol resistance. | ||
| + | Thus, if a cell has taken up the vector which is encoding the gene of interest as well as the resistance gene this cell | ||
| + | becomes resistant against chloramphenicol. | ||
| + | |||
| + | <h2>Resistance</h2> | ||
| + | |||
| + | <div class="half right"> | ||
| + | <figure class="figure large"> | ||
| + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/8/80/T--Bielefeld-CeBiTec--Chloramphenicol_Acetyltransferase_NMR.png" alt=""> | ||
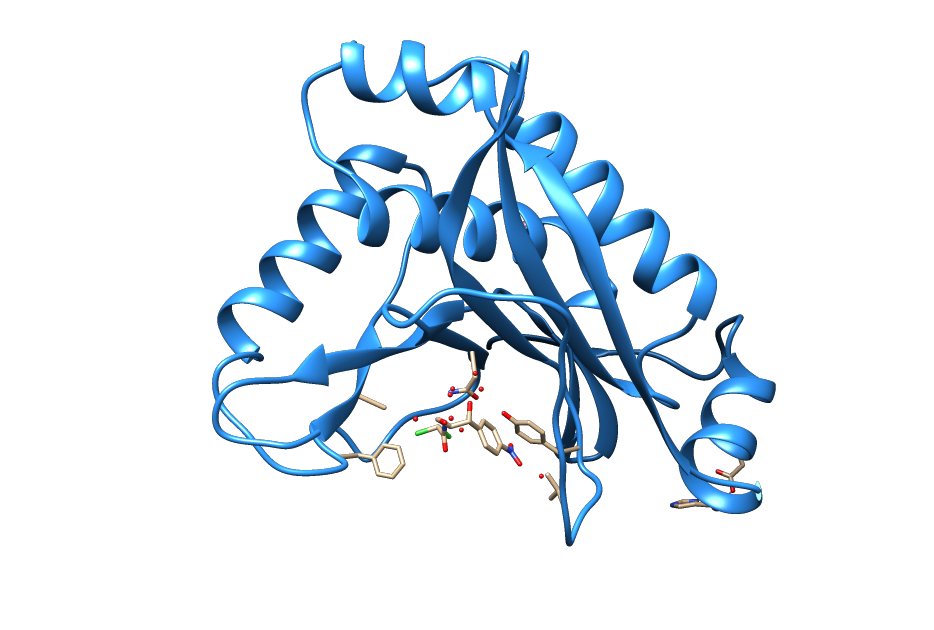
| + | <figcaption>Ribbon structure of the chloramphenicol acetyltransferase chloramphenicol bound to the catalytic center. (From PDB: 3CLA)(Leslie, A.G.W. 1990)</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | The choloramphenicol acetyltransferase (CAT) is an enzyme originally identified in <i>Escherichia coli</i>, that mediates resistance to chloramphenicol. (Shaw 1983) CAT covalently attaches an acetyl group from acetyl-CoA to chloramphenicol, rendering the acetylated chloramphenicol unable to bind to the 23S rRNA. (Shaw 1991) | ||
| + | It is encoded by the <i>cat</i> gene which is commonly used by the iGEM community as part <a href=”https://parts.igem.org/Part:BBa_J31005”>BBa_J31005</a> and in the standard backbone <a href="https://parts.igem.org/Part:pSB1C3:Design">pSB1C3</a>. | ||
| + | <!-- <h2>Economical advantages</h2> --> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <a class="anchor" id="h2"></a> | ||
| + | <h1 id="a2">Intein-mediated protein splicing. </h1> | ||
| + | <hr> | ||
| + | |||
| + | |||
| + | <h2>Inteins</h2> | ||
| + | |||
| + | <div class="middle"> | ||
| + | <figure class="figure large"> | ||
| + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/e/e7/T--Bielefeld-CeBiTec--basics_Intein.jpg" alt=""> | ||
| + | <figcaption>Mechanism of intein-mediated protein splicing.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | An intein is a segment of a precursor protein which is able to mediate its own excision form the precursor protein, while joining the flanking protein sequences together, creating a new peptide bond. <br> | ||
| + | The intein-mediated protein splicing occurs after the mRNA is translated into a sequence of amino acids. Prior to the splicing process the N-terminal part of the precursor protein is called N-extein, the center part is the intein and the C-terminal part is named C-extein. The spliced protein is called an extein as well. <br> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <h2>Split inteins</h2> | ||
| + | A very special but small subset of inteins are the so-called split-inteins. They are transcribed and translated as separate polypeptides but rapidly associate afterwards to form the active intein. The active intein then ligates the fused N- and C-exteins in a process called protein trans-splicing. | ||
| + | |||
| + | </div> | ||
| + | <div class="middle"> | ||
| + | <figure style="width:400px" class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/8/88/T--Bielefeld-CeBiTec--basics_splitintein.png" alt=""> | ||
| + | <figcaption>Mechanism of split intein-mediated protein splicing. The intein autocatalytically cleaves itself out of the polypeptide, connecting both adjacent exteins by a peptide bond. </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <div> | ||
| + | |||
| + | After the first split intein SspDnaE, a subunit of the DNA polymerase III (DnaE) from <i>Synechocystis sp. strain</i> PCC6803 was discovered. Several homologous inteins were identified in cyanobacteria. (Wei et al. 2006, Dassa et al. 2007) However apart from sequence analysis there have only been few attempts to further characterize or compare their splicing efficiencies. (Dassa et al. 2007) Nevertheless a split intein from the cyanobacterium <i>Nostoc punctiforme</i> (NpuDnaE) which is highly homologous to Ssp DnaE was analyzed quite extensively. It has shown higher activities than SspDnaE <i>in vivo</i> and <i>in vitro</i>. (Iwai et al. 2006; Zettler et al. 2009). Additionally it has a boarder range of acceptance when it comes to the residues at the end of the C-extein. (Iwai et al. 2006).<br> | ||
| + | <!-- which is a subunit of the DNA polymerase III (DnaE) from <i>Nostoc punctiforme</i>. | ||
| + | however, Npu DnaE can splice a bride range of sequences, due to the variable extein types. Some result in faster splice rates compared to the native ones (Cheriyan et al., 2013). --> | ||
| + | </div> | ||
| + | |||
| + | <h2>Amino acids neighboring split sites </h2> | ||
| + | |||
| + | <div class="half right"> | ||
| + | <figure class="figure large"> | ||
| + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/4/47/T--Bielefeld-CeBiTec--basicpart_Intein3d.png" alt=""> | ||
| + | <figcaption>NMR solution structure of NpuDnaE (PDB code: 2KEQ) (Oeemig et al. 2009)</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
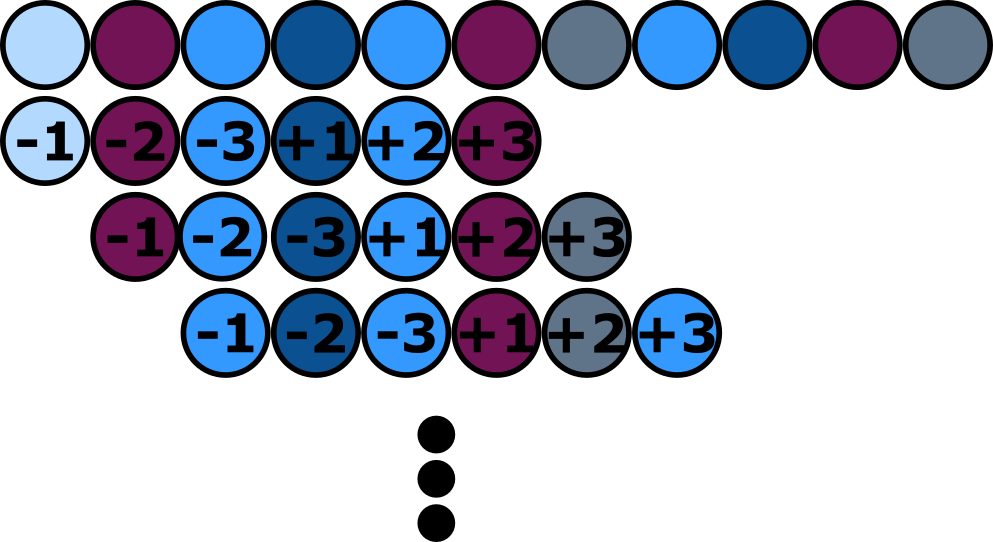
| + | As implied, the activity of most split inteins is strongly dependent on the residues of the amino acids at the transition points between the intein and the extein. Therefore, the number of ligation sites for a natural split intein within a target protein is limited to sequences similar to endogenous extein sequences. The intein autocatalytically cleaves itself out of the polypeptide, connecting both adjacent exteins by a peptide bond. It has been shown that some amino acids are more favorable in the positions neighboring the splicing site than others. Especially the three amino acids right before (in the N-extein) and right after (in the C-extein) the splicing site are crucial. Not only the likelihood of correct splicing but also the speed of the splicing reaction are strongly depending on these amino acids. In almost all cases cytosin must be in the +1 position of the C-extein. For more information about the other amino acids possible in the crucial positions have a look at our <a href="https://2019.igem.org/Team:Bielefeld-CeBiTec/Model#h1">Model page</a>. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="center"> | ||
| + | <figure class="figure large"> | ||
| + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/0/04/T--Bielefeld-CeBiTec--Split_Neighb.png"> | ||
| + | <figcaption>The highlighted three amino acids in the exteins bordering on the inteins are the most relevant for a successful splicing process. These are the amino acids are the ones that are known to have the biggest impact on the functionality of the splicing process (Cheriyan, Pedamallu, Tori, & Perler, 2013). | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Mechanism</h2> | ||
| + | |||
| + | For a better understanding of the importance of the amino acid residues near the splicing site, following is a short summary of the split-intein-mediated splicing mechanism:: | ||
| + | |||
| + | |||
| + | <div class=""> | ||
| + | <figure class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/6/6a/T--Bielefeld-CeBiTec--Intein_Splicing.png" alt=""> | ||
| + | <figcaption>Intein-mediated splicing mechanism</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <ol> | ||
| + | <li> Split inteins associate</li> | ||
| + | <li> Nucleophilic attack of the peptide bond between the N-extein and the intein by the residue of the first amino acid in the intein</li> | ||
| + | <li> Formation of a linear ester (not shown) or thioester intermediate </li> | ||
| + | <li> Side chain of the first residue of the C-extein attacks the thioester (or ester), freeing the N-terminal end of the intein</li> | ||
| + | <li> N- and C-extein are connected, but not yet by a peptide bond</li> | ||
| + | <li> Amide nitrogen atom from the residue of the last amino acid of the intein cleaves the peptide bond between the intein and the C-extein</li> | ||
| + | <li> Intein is free</li> | ||
| + | <li> Free amino group of the C-extein attacks thioester (or ester) linking the exteins together </li> | ||
| + | <li> O-N or S-N shift forms a peptide bond creating a fully functional protein</li> | ||
| + | </ol> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <a class="anchor" id="h3"></a> | ||
| + | <h1 id="a3">Split-resistance genes</h1> | ||
| + | <hr> | ||
| + | |||
| + | Split inteins enable us to divide and fuse all kinds of proteins. | ||
| + | Their application ranges from structural biology, especially NMR spectroscopy to split intein-mediated production | ||
| + | of genetically encoded cyclic peptide libraries. (Horshill and Benkovic 2006; Volkmann and Iwai 2010) | ||
| + | Another possible application are split markers, like antibiotics for the selection of GMOs. <br> | ||
| + | Researchers working in the field of synthetic biology are often using two or multiple plasmid systems to incorporate | ||
| + | all desired transgenes into one cell. For the selection of positive clones and stable retention of the plasmids they usually apply several antibiotics to the | ||
| + | cultivation medium. Unfortunately, multiple antibiotics provide very harsh growth conditions for the cells. <br> | ||
| + | Split antibiotics are a great solution to reduce the number of needed antibiotics. A scheme how split markers are | ||
| + | implemented is illustrated in figure 8. To create a split selectable marker the gene encoding an antibiotic resistance is | ||
| + | split into two segments. Each segment is fused to one part of a split intein which is able to join the protein segments back together. Each of these fusion segments is inserted into a different vector. If a cell is transformed with both cohesive vectors it is able to develop a resistance against the respective antibiotic via protein trans-splicing. | ||
| + | If the cell has taken up only one of the vectors it has no chance to survive selection since it can produce only one part | ||
| + | of the protein mediating the antibiotic resistance. <br> | ||
| + | |||
| + | |||
| + | <div class="center"> | ||
| + | <figure class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/d/d6/T--Bielefeld-CeBiTec--split_resistance.jpg"> | ||
| + | <figcaption> Mechanism of split resistance genes | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Using split selectable markers it is possible keep two vectors encoding genes of interest inside a cell while using only | ||
| + | one instead of two antibiotics. Which is beneficial not only for the growth conditions of the cells but also for the financial | ||
| + | aspects of a project, since antibiotics are relatively expensive. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <a class="anchor" id="h4"></a> | ||
| + | <h1 id="a4">Biosafety</h1> | ||
| + | <hr> | ||
| + | When using split selectable markers one part of the protein is encoded by a plasmid whereas the other part could be | ||
| + | encoded by either another plasmid or the genome of the target organism. | ||
| + | In both cases this system would increase the biosafety of the systems encoded on the plasmids. If one of the plasmids, | ||
| + | a strain containing only one of the plasmids, or a strain encoding one of the parts in its genome would be released into | ||
| + | the environment the antibiotic resistance would not spread. Since both parts of the protein are needed to establish the | ||
| + | resistance, the uptake of only one of the parts would not increase the fitness of an organism in the outer world. | ||
| + | This reduces the probability of GMOs to overgrow natural populations in the environment. | ||
| + | Therefore, this system could increase the safety of GMOs in any application outside of the lab. | ||
| + | This would be beneficial for the future application of many iGEM projects. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a class="anchor" id="h5"></a> | ||
| + | <h1 id="a5">Model and plasmid design</h1> | ||
| + | <hr> | ||
| + | |||
| + | <h2>Preliminary considerations</h2> | ||
| + | |||
| + | To assemble our Troygenics we are working with a two plasmid system. | ||
| + | Since most of the real world applications of the Troygenics involve their release into the outer world, | ||
| + | the biosafety of our system is a crucial part of our project. | ||
| + | As explained above split selectable markers are a great way to improve the biosafety of two plasmid systems, | ||
| + | because they not only reduce the likelihood of spread antibiotic resistances but also reduce the probability of | ||
| + | GMOs overgrowing naturally occurring populations. | ||
| + | |||
| + | As chloramphenicol is the standard antibiotic resistance used by the iGEM community | ||
| + | but was never submitted as a split antibiotic resistance before we decided to | ||
| + | develop a split chloramphenicol resistance, which is particular useful for future iGEM teams. | ||
| + | |||
| + | |||
| + | It was our aim to split the resistance gene cat so that the two parts of the chloramphenicol acetytransferase are | ||
| + | encoded by two different plasmids. According to the advantages mentioned above, we decided to use the split | ||
| + | intein NpuDnaE (Partnummern)to create our kanamycin split resistance. <br> | ||
| + | The important amino acids neighboring the native splice site of NpuDnaE are“Ala Glut Tyr” for the N-extein and | ||
| + | “Cys Phe Asn” for the native C-extein. (Cheriyan et al. 2013) To examine the optimal split point of the chloramphenicol | ||
| + | acetyltransferase which mediates the chloramphenicol resistance we designed and executed a <a href="https://2019.igem.org/Team:Bielefeld-CeBiTec/Model">model</a>. | ||
| + | It provides us with a list of all possible split points, ranked from best to worst, for the desired protein, based on its amino acid sequence and 3D structure. | ||
| + | |||
| + | <h2>Modeling the optimal split point </h2> | ||
| + | <div> | ||
| + | Our model takes the structure as well as the sequences around potential split points into account. At first split-points in regions which form an alpha-helix or a beta-sheet are avoided. Additionally the amino acids neighboring the splicing site are inspected. | ||
| + | As previously shown, that certain amino acids are more favorable in these positions, leading to more correct splicing events as to an overall increased splicing rate. | ||
| + | Based on previously reported data (Cheriyan, Pedamallu, Tori, & Perler, 2013), a score between 0 and 50 has been assigned to imply the value of an amino acid at a certain position. The higher the value the more favorable is the amino acid in that position for faster and more specific splicint. | ||
| + | The positions relevant for this kind of splicing are the three ones right before and right after the insertion site for the split-intein (Fig. 1). | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | The <a href="https://2019.igem.org/Team:Bielefeld-CeBiTec/Model">modeling</a> results indicate that the best split point | ||
| + | would be after amino acid 30. | ||
| + | <div class="middle"> | ||
| + | <table> | ||
| + | <caption> Relative favorability of each amino acid i (rF(A<sub>i</sub>)) at each relevant position j neighboring the inteins. Adjusted from Cheriyan, Pedamallu, Tori, & Perler, 2013. The amino acids neighboring a splicing site were randomly mutated, +1 was set to Cysteine. The mutants conducting successful protein splicing were selected and the 6 neighboring amino acids were determined. To obtain the rf(A<sub>i</sub>) values the occurence of each amino acid in each position was counted and this number was divided by the natural frequency of said amino acid. For our model Cysteine +1 was set to “50” to ensure that this was picked over any other combinations. Additionally, Methionine +2 was set to 20, as other sources stated that it might be an appropriate substitute for Cysteine (Brenzel, 2009). </caption> | ||
| + | <thead id="tablehead"> | ||
| + | <tr> | ||
| + | <th style="width: auto;"> Amino Acid </th> | ||
| + | <th colspan="3" style="width: auto;"> rF(A<sub>i</sub>) at the N-terminal extein</th> | ||
| + | <th colspan="3" style="width: auto;"> rF(A<sub>i</sub>) at the C-terminal extein </th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <thead id="tablehead"> | ||
| + | <tr> | ||
| + | <th style="width: auto;"> </th> | ||
| + | <th style="width: auto;"> -3 </th> | ||
| + | <th style="width: auto;"> -2 </th> | ||
| + | <th style="width: auto;"> -1 </th> | ||
| + | <th style="width: auto;"> +1 </th> | ||
| + | <th style="width: auto;"> +2 </th> | ||
| + | <th style="width: auto;"> +3 </th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> D </th> | ||
| + | <td style="width: auto;"> 1.39 </th> | ||
| + | <td style="width: auto;"> 1.52 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> E </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | <td style="width: auto;"> 3.51 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 5.5 </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> N </th> | ||
| + | <td style="width: auto;"> 0.93 </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | <td style="width: auto;"> 2.19 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 8.61 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> Q </th> | ||
| + | <td style="width: auto;"> 1.39</th> | ||
| + | <td style="width: auto;"> 0.86 </th> | ||
| + | <td style="width: auto;"> 0.33 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.27 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> H </th> | ||
| + | <td style="width: auto;"> 1.06 </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | <td style="width: auto;"> 2.19 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 8.61 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;">K </th> | ||
| + | <td style="width: auto;"> 1.59 </th> | ||
| + | <td style="width: auto;"> 0.93 </th> | ||
| + | <td style="width: auto;"> 4.44 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> R </th> | ||
| + | <td style="width: auto;"> 1.81</th> | ||
| + | <td style="width: auto;"> 0.2 </th> | ||
| + | <td style="width: auto;"> 0.86 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> S </th> | ||
| + | <td style="width: auto;"> 0.99</th> | ||
| + | <td style="width: auto;"> 1.24 </th> | ||
| + | <td style="width: auto;"> 1.37 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> C </th> | ||
| + | <td style="width: auto;"> 0.46</th> | ||
| + | <td style="width: auto;"> 0.40</th> | ||
| + | <td style="width: auto;"> 0.80 </th> | ||
| + | <td style="width: auto;"> 50 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> T </th> | ||
| + | <td style="width: auto;"> 0.63</th> | ||
| + | <td style="width: auto;"> 0.89 </th> | ||
| + | <td style="width: auto;"> 1.59 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.96 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> P </th> | ||
| + | <td style="width: auto;"> 0.80 </th> | ||
| + | <td style="width: auto;"> 1.52 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> G </th> | ||
| + | <td style="width: auto;"> 4.67 </th> | ||
| + | <td style="width: auto;"> 2.68 </th> | ||
| + | <td style="width: auto;"> 0.17 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.03 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> A </th> | ||
| + | <td style="width: auto;"> 0.63 </th> | ||
| + | <td style="width: auto;"> 0.83 </th> | ||
| + | <td style="width: auto;"> 1.92 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> V </th> | ||
| + | <td style="width: auto;"> 0.56 </th> | ||
| + | <td style="width: auto;"> 0.50 </th> | ||
| + | <td style="width: auto;"> 0.23 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.03 </th> | ||
| + | <td style="width: auto;"> 0.10 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> I </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 0.53 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.40 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> L </th> | ||
| + | <td style="width: auto;"> 0.11 </th> | ||
| + | <td style="width: auto;"> 0.57 </th> | ||
| + | <td style="width: auto;"> 0.75 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.82 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> M </th> | ||
| + | <td style="width: auto;"> 0.13 </th> | ||
| + | <td style="width: auto;"> 0.73 </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | <td style="width: auto;"> 20 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 2.25 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> F </th> | ||
| + | <td style="width: auto;"> 0.13 </th> | ||
| + | <td style="width: auto;"> 0.73 </th> | ||
| + | <td style="width: auto;"> 1.26 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 2.25 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> Y </th> | ||
| + | <td style="width: auto;"> 0.20 </th> | ||
| + | <td style="width: auto;"> 0.60 </th> | ||
| + | <td style="width: auto;"> 2.05 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 0.13 </th> | ||
| + | <td style="width: auto;"> 5.23 </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> W </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | <td style="width: auto;"> 0.40 </th> | ||
| + | <td style="width: auto;"> 0.99 </th> | ||
| + | <td style="width: auto;"> 0.00 </th> | ||
| + | <td style="width: auto;"> 29.42 </th> | ||
| + | <td style="width: auto;"> 0.07 </th> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | <div class=""> | ||
| + | To identify the optimal split point for intein-mediated splicing of proteins, all possible 6 amino acid long fragments of the peptide sequences were assessed by a python script. | ||
| + | The sum of their rf(A<sub>i</sub>)<sub>j</sub> values was calculated for each fragment, revealing how well the center of the respective fragment would act as a split point. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="middle"> | ||
| + | <figure class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/7/73/T--Bielefeld-CeBiTec--Seperation_Proteins.png"> | ||
| + | <figcaption>The generation of all possible 6 amino acid long fragments of the peptide sequence of interest. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class=""> | ||
| + | <br> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="middle"> | ||
| + | <figure style="width:400px" class="figure large"> | ||
| + | <img style="height:10vh" class="figure image" src="https://2019.igem.org/wiki/images/d/dd/T--Bielefeld-CeBiTec--Formula_BenSum.png"> | ||
| + | <figcaption> <b> Formula used to calculate how beneficial a certain combination of amino acids is for Npu DnaE mediated protein splicing. </b> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class=""> | ||
| + | <br> | ||
| + | Using this approach, a list with all possible split points sorted by B(seq)<sub>n</sub> is created, ranging from the best to the worst possible split point in the protein. <br> | ||
| + | Complementing the sequence based determination of potential split points the desired structure of the final protein has to be considered. Positions in important structural features of the protein were determined using the protein structure viewer Chimera (Pettersen et al., 2004). Sequences involved in relevant structures were integrated in the analyses as described above to prevent the identification of possible split points in these regions. | ||
| + | Lastly, the remaining list of split points was sorted from best to worst and the best split sequences determined were written FASTA files. Using the MODELLER-software (Sali & Webb, 1989), the most likely structure of the N- or C-terminal part of the protein fused to the split intein, prior to finding the other intein in the cell, was determined. To do so, it was necessary to find proteins with similar structures as templates for homology modeling. They were chosen as follows: </div> | ||
| + | |||
| + | <div class=""> | ||
| + | <br> | ||
| + | |||
| + | <a class="anchor" id="h3"></a> | ||
| + | <h1 id="a3"> Split Chloramphenicol Resistance</h1> | ||
| + | <hr> <!--Blaue Linie--> | ||
| + | <div class=""> | ||
| + | We aimed to split the protein conveying resistance to chloramphenicol, chloramphenicol acetyltransferase (BBa_J3105). The resistance protein is commonly used within iGEM, thus making a split Chloramphenicol acetyltransferasa a vluable addition to the iGEM partsreg. The protein sequence of this part can be found <a href="https://www.uniprot.org/uniprot/P62577">here</a>. | ||
| + | According to our modeling, the best split point to use when introducing Npu DnaE was VAQ-CTY in the positions 28-33, the B(<sub>VAQCTY</sub>) is 56.95.<br> | ||
| + | Even though this split point is located at the second amino acid of a beta-sheet, we decided to test whether splitting at this position could lead to Chloramphenicol acetyltransferase fragments that could be reconstituted by intein-mediated protein splicing. <br> | ||
| + | |||
| + | Similar to our modeling of the split kanamycin resistance, we used the homology modeling software MODELLER (Sali & Webb, 1989) to visualize each part of the chloramphenicol acetyltransferase fused to the Npu DnaE intein. <br> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | |||
| + | The amino acid sequences given to the software were: | ||
| + | </div> | ||
| + | |||
| + | <div id="rcorners1" style="word-wrap: break-word;" class=""> | ||
| + | Part 1: <br> | ||
| + | <font color="#39f"> MEKKITGYTTVDISQWHRKEHFEAFQSVAQ</font><font color="#800000">CLSYETEILTVEYGLLPIGKIVEKRIECTVYSVDNNGNIYTQPVAQWHDRGEQEVFEYCLEDGSLIRATKDHKFMTVDGQMLPIDEIFERELDLMRVDNLPNNLEGHHHHHH</font><br><br> | ||
| + | |||
| + | Part 2: <br> | ||
| + | <font color="#800000">MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNC</font><font color="#39f">CTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA</font> | ||
| + | </div> | ||
| + | |||
| + | <div class=""> | ||
| + | <br>The templates used for homology modeling were:<br><br> | ||
| + | <div class="middle"> | ||
| + | <table> | ||
| + | </thead> | ||
| + | <thead id="tablehead"> | ||
| + | <tr> | ||
| + | <th style="width: auto;"> Part 1 </th> | ||
| + | <th style="width: auto;"> Part 2 </th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> 1NOC, chain B (Chloramphenicol acetyltransferase) </th> | ||
| + | <td style="width: auto;"> 1NOC, chain B (Chloramphenicol acetyltransferase) </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> 4QFQ, chain A (Npu DnaE) </th> | ||
| + | <td style="width: auto;"> 4QFQ, chain B (Npu DnaE) </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="width: auto;"> 5OL6, chain A (inactivated Npu SICLOPPS intein with CAFHPQ extein) </th> | ||
| + | <td style="width: auto;"> 5OL6, chain B (inactivated Npu SICLOPPS intein with CAFHPQ extein) </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> 4KL5, chain A (Npu DnaE)</th> | ||
| + | <td style="width: auto;"> 1QCA, chain A (Type III Chloramphenicol acetyltransferase) </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto;"> 2KEQ, chain A (DnaE intein from Nostoc punctiforme) </th> | ||
| + | <td style="width: auto;"> 3CLA, chain A (Type III Chloramphenicosl acetyltransferase)</th> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | After running the MODELLER software (Sali & Webb, 1989), two protein structures were calculated and placed adjacent to each other using Chimera (Pettersen et al., 2004). | ||
| + | |||
| + | |||
| + | |||
| + | <div class=""> | ||
| + | <figure style="width:400px" class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/0/02/T--Bielefeld-CeBiTec--Cm_Split_VAQCTY.png"> | ||
| + | <figcaption> The predicted 3D-structure of each subunit of the Cloramphenicol acetyltransferase split at the optimal predicted split point VAQ-CTY prior to splicing. It was developed using the homology modeling software MODELLER (Sali & Webb, 1989). | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <h2>Plasmid design</h2> | ||
| + | |||
| + | We designed 4 parts encoding this chloramphenicol split resistance. | ||
| + | Part <a href="https://parts.igem.org/Part:BBa_K2926076">BBa_2926076</a> and <a href="https://parts.igem.org/Part:BBa_K2926077">BBa_2926076</a> are basic parts. | ||
| + | <a href="https://parts.igem.org/Part:BBa_K2926076">BBa_2926076</a> is encoding the N-terminal part of the split resistance, fused to the N-intein. This part was constructed in <a href="https://parts.igem.org/Part:pSB3T5">pSB3T5</a>. | ||
| + | Whereas <a href="https://parts.igem.org/Part:BBa_K2926077">BBa_2926076</a> encodes the C-intein fused to the C-terminal part of the split resistance and was constructed in <a href="https://parts.igem.org/Part:pSB1K3">pSB1K3</a>.<br> | ||
| + | |||
| + | |||
| + | Additionally to the split chloramphenicol resistance described on this page, we have constructed another chloramphenicol split resistance, | ||
| + | as well as a kanamycin, ampicilin and a hygromycin | ||
| + | split resistance. <a href="https://2019.igem.org/Team:Bielefeld-CeBiTec/Part_Collection">Here</a> you can get more information on our | ||
| + | handy <a href="https://2019.igem.org/Team:Bielefeld-CeBiTec/Part_Collection">collection of split antibiotic resistances</a>. | ||
| + | |||
| + | </div> | ||
| + | <div> | ||
| + | |||
| + | <a class="anchor" id="h6"></a> | ||
| + | <h1 id="a6">Characterization</h1> | ||
| + | <hr> | ||
| + | |||
| + | We have successfully constructed all parts and verified their nucleotide sequence with sanger sequencing.<br> | ||
| + | After co-transformation of <a href="https://parts.igem.org/Part:BBa_K2926076">BBa_2926076</a> and <a href="https://parts.igem.org/Part:BBa_K2926077">BBa_2926076</a> into <i>E. coli DH5α</i> the cells where plated on selection plates containing kanamycin and tetracyclin, but no chloramphenicol | ||
| + | since the cells need some time to establish the chloramphenicol resistance. <br> | ||
| + | Afterwards a number of colonies were transferred to numerous plates, each containing a different concentration from 0.1 up to 100 μg/mL chloramphenicol. | ||
| + | Alongside with our colonies containing the split chloramphenicol resistance we plated <i>E. coli DH5α</i> as a | ||
| + | negativ control and <i>E. coli DH5α</i> containing pSB1C3, which is encoding RFP as a positive control.<br> | ||
| + | |||
| + | We conducted this experiment as a triplicate. The results of one of the triplicates are depicted in figure 10. | ||
| + | Native <i>E. coli DH5α</i> was only growing on the plates containing 0.1 μg/mL chloramphenicol, implying that the native <i>E. coli DH5α</i> has no significant resistance to chloramphenicol. | ||
| + | <i>E. coli DHα</i> containing pSB1C3, which is encoding RFP was proven to be resistant on all tested chloramphenicol concentrations. | ||
| + | The <i>E. coli DH5α</i> encoding our split chloramphenicol resistance showed to be resistant to chloramphenicol concentrations | ||
| + | of averaged 7 μg/mL.<br> | ||
| + | |||
| + | In every day lab life chloramphenicol is used in concentrations between 10 and 25 μg/mL. | ||
| + | Since our split system shows resistance against chloramphenicol for concentrations of up to averaged 7 μg/mL | ||
| + | the working concentrations for chloramphenicol could be reduced, | ||
| + | leading to a diminished chloramphenicol consumption. | ||
| + | |||
| + | <div class=""> | ||
| + | <figure style="width:400px" class="figure large"> | ||
| + | <img class="figure image" src="https://2019.igem.org/wiki/images/0/08/T--Bielefeld-CeBiTec--Split_Cm_A_B.png"> | ||
| + | <figcaption><b>Resistance test of split chloramphenicol resistance</b> with | ||
| + | <i>E. coli DH5α</i> (in the top left section of every plate), | ||
| + | <i>E. coli DH5α</i> containing pSB1C3_RFP (in the bottom section of every plate) and | ||
| + | <i>E. coli DH5α</i> containing our chloramphenicol split resistance (in the top right section of every plate) | ||
| + | plated on LB plates with of 0.1, 1, 7, 10, 20, 40, 60, 80, 100 μg/mL chloramphenicol (left to right and top to bottom) | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <h1>List of our basic Parts</h1> | ||
| + | <hr> | ||
| + | <div> | ||
| + | <table> | ||
| + | <caption> List of all basic parts </caption> | ||
| + | <thead id="tablehead"> | ||
| + | <tr> | ||
| + | <th style="width: 20%"> Basic parts </th> | ||
| + | <th style="width: 30%!important"> Description </th> | ||
| + | <th style="width: 22,5%"> Designer </th> | ||
| + | <th style="width: auto"> Length </th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926000"target="_blank">BBa_K2926000</a> </td> | ||
| + | <td style="width: auto"> Cas13a Lsh</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 4168 bp </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926001 | ||
| + | "target="_blank">BBa_K2926001</a> </td> | ||
| + | <td style="width: auto"> Cas13a Lbu</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 3480 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr><td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926003 | ||
| + | "target="_blank">BBa_K2926003</a> </td> | ||
| + | <td style="width: auto"> GALL yeast promoter (validated part)</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 430 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr><td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926004 | ||
| + | "target="_blank">BBa_K2926004</a> </td> | ||
| + | <td style="width: auto"> SNR52 yeast promoter</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 269 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926005"target="_blank">BBa_K2926005</a> </td> | ||
| + | <td style="width: auto"> TPS1 yeast terminator</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 250 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926006"target="_blank">BBa_K2926006</a> </td> | ||
| + | <td style="width: auto"> SUP4 yeast terminator</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 20 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926007"target="_blank">BBa_K2926007</a> </td> | ||
| + | <td style="width: auto"> Inverted terminal repeat form S. cerevisiae gen ADE2, front</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 50 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926008"target="_blank">BBa_K2926008</a> </td> | ||
| + | <td style="width: auto"> gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA1-3</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 187 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926009"target="_blank">BBa_K2926009</a> </td> | ||
| + | <td style="width: auto"> gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA1-7</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 435 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926010"target="_blank">BBa_K2926010</a> </td> | ||
| + | <td style="width: auto"> gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA4-7</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 301 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926011"target="_blank">BBa_K2926011</a> </td> | ||
| + | <td style="width: auto"> gRNA array (cas13a) with 3 sgRNAs (for A. niger)</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 207 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926013"target="_blank">BBa_K2926013</a> </td> | ||
| + | <td style="width: auto"> Inverted terminal repeat form S. cerevisiae gen ADE2, back</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 50 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926015"target="_blank">BBa_K2926015</a> </td> | ||
| + | <td style="width: auto">Aspergillus niger 5S rRNA</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 119 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926020"target="_blank">BBa_K2926020</a> </td> | ||
| + | <td style="width: auto">Promotor of protein VIII from the M13 phage</td> | ||
| + | <td style="width: auto"> Astrid Többer</td> | ||
| + | <td> 47 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926021"target="_blank">BBa_K2926021</a> </td> | ||
| + | <td style="width: auto">sgRNA for the detection of RFP with Cas13a</td> | ||
| + | <td style="width: auto"> Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth </td> | ||
| + | <td> 57 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926022"target="_blank">BBa_K2926022</a> </td> | ||
| + | <td style="width: auto">Trp_sfGFP_Opy2p_mCherry_M13K07 gene VIII_M13K07 gene III</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 4066 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926023"target="_blank">BBa_K2926023</a> </td> | ||
| + | <td style="width: auto">M13K07_genes_II_VIII</td> | ||
| + | <td style="width: auto"> Astrid Többer </td> | ||
| + | <td> 2015 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926025"target="_blank">BBa_K2926025</a> </td> | ||
| + | <td style="width: auto">M13K07_genes_III_IV</td> | ||
| + | <td style="width: auto"> Astrid Többer </td> | ||
| + | <td> 2820 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926026"target="_blank">BBa_K2926026</a> </td> | ||
| + | <td style="width: auto">M13K07_truncated_III</td> | ||
| + | <td style="width: auto"> Astrid Többer </td> | ||
| + | <td> 456 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926027"target="_blank">BBa_K2926027</a> </td> | ||
| + | <td style="width: auto">mCherry_VIII_fusion_protein</td> | ||
| + | <td style="width: auto"> Astrid Többer </td> | ||
| + | <td> 1086 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926048"target="_blank">BBa_K2926048</a> </td> | ||
| + | <td style="width: auto">mCherry with hexahistidine tag for purification</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 729 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926049"target="_blank">BBa_K2926049</a> </td> | ||
| + | <td style="width: auto">Mating factor alpha from S. cerevisiae fused to mCherry</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 762 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926050"target="_blank">BBa_K2926050</a> </td> | ||
| + | <td style="width: auto">Fusion protein of Flo11 from yeast and Cherry</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1287 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926051"target="_blank">BBa_K2926051</a> </td> | ||
| + | <td style="width: auto">Fusion protein of Opy2 from yeast and Cherry</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 837 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926052"target="_blank">BBa_K2926052</a> </td> | ||
| + | <td style="width: auto">Fusion protein of mCherry and M13 bacteriophage protein pVIII</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 942 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926053"target="_blank">BBa_K2926053</a> </td> | ||
| + | <td style="width: auto">Fluorescence reporter mCherry fused to pVIII from M13 bacteriophage with purification tag</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 729 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926054"target="_blank">BBa_K2926054</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of mating factor alpha from yeast, mCherry, and pVIII from M13 bacteriophage</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 994 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926055"target="_blank">BBa_K2926055</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of Flo11 from yeast, mCherry, and pVIII from M13 bacteriophage</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1521 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926056"target="_blank">BBa_K2926056</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of Opy2 from yeast, mCherry, and pVIII from M13 bacteriophage</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1071 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926057"target="_blank">BBa_K2926057</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of mating factor alpha, mCherry, and pVIII with His-tag</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1014 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926058"target="_blank">BBa_K2926058</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of Flo11 (yeast), mCherry, and pVIII (M13 bacteriophage) with purification tag</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1539 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926067"target="_blank">BBa_K2926067</a> </td> | ||
| + | <td style="width: auto"> Fusion protein of Opy2 (yeast), mCherry and pVIII (bacteriophage M13) with purification tag</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 1089 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926068"target="_blank">BBa_K2926068</a> </td> | ||
| + | <td style="width: auto"> Proline-glycine-peptide fused to the n-terminus of mCherry</td> | ||
| + | <td style="width: auto"> Johanna Opgenoorth </td> | ||
| + | <td> 750 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926076"target="_blank">BBa_K2926076</a> </td> | ||
| + | <td style="width: auto"> Beta-Cmp resistance protein part 1 (Cm_B_split1_NpuN_pSB3Cd5_Kan)/td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 537 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926077"target="_blank">BBa_K2926077</a> </td> | ||
| + | <td style="width: auto"> Beta-Cmp resistance protein part 2 (NpuC_Cm_B_split2_pSB1Cc3_Tet)/td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 543 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926078"target="_blank">BBa_K2926078</a> </td> | ||
| + | <td style="width: auto"> Alpha-Kan resistance protein part 1 (Kan_A_split1_NpuN_pSB3Kb5_Cm)</td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 591 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926079"target="_blank">BBa_K2926079</a> </td> | ||
| + | <td style="width: auto"> Alpha-Kan resistance protein part 2 (NpuC_Kan_A_split2_pSB1Ka3_Tet)</td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 645 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926080"target="_blank">BBa_K2926080</a> </td> | ||
| + | <td style="width: auto"> Alpha-Amp resistance protein part 1 (Amp_A_Split1_NpuN_pSB1Aa3_Tet)</td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 645 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="width: auto"> <a href="https://parts.igem.org/Part:BBa_K2926081"target="_blank">BBa_K2926081</a> </td> | ||
| + | <td style="width: auto"> Alpha-Amp resistance protein part 2 (NpuC_Amp_A_split2_pSB3Ab5_Cm)</td> | ||
| + | <td style="width: auto"> Nefeli Chanoutsi </td> | ||
| + | <td> 645 bp </td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | <div id="Ende"> </div> <!--For the processbar please do not delete--> | ||
| + | |||
| + | <details> | ||
| + | <summary> References </summary> | ||
| + | <p> Shah, Neel H.; Muir, Tom W. (2011): Split Inteins: Nature's Protein Ligases. In: Israel Journal of Chemistry 51 (8-9). <a href="https://doi.org/10.1002/ijch.201100094">Link</a> </p> | ||
| + | <p> Zettler, Joachim; Schütz, Vivien; Mootz, Henning D. (2009): The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. In: FEBS Letters 583. <a href="https://doi.org/10.1016/j.febslet.2009.02.003">Link</a> </p> | ||
| + | <p> Jillette, Nathaniel; Du, Menghan; Cheng, Albert (2019): Split Selectable Markers. In: bioRxiv. <a href="http://dx.doi.org/10.1101/452979"> Link</a> </p> | ||
| + | <p> Iwai, Hideo; Züger, Sara; Jin, Jennifer; Tam, Pui-Hang (2006): Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. In: FEBS Letters 580 (7). <a href="https://doi.org/10.1016/j.febslet.2006.02.045">Link</a></p> | ||
| + | <p> Cheriyan, Manoj; Pedamallu, Chandra S.; Tori, Kazuo; Perler, Francine (2013): Faster Protein Splicing with the Nostoc punctiforme DnaE Intein Using Non-native Extein Residues. In: Journal of Biological Chemistry 288 (6). <a href="http://www.jbc.org/content/288/9/6202"> Link</a></p> | ||
| + | <p> Oeemig, Jesper S.; Aranko, A. Sesilja; Djupsjöbacka, Janica; Heinämäki, Kimmo; Iwai, Hido (2009): Solution structure of DnaE intein from Nostoc punctiforme: Structural basis for the design of a new split intein suitable for site‐specific chemical modification. In: FEBS Letters 583 (9). <a href="https://doi.org/10.1016/j.febslet.2009.03.058">Link</a> </p> | ||
| + | <p> Dassa, Bareket; Amitai, Gil; Caspi, Jonathan; Schueler-Furman, Ora; Pietrokovski, Shmuel (2007): Trans Protein Splicing of Cyanobacterial Split Inteins in Endogenous and Exogenous Combinations. In: Biochemistry 46 (1). <a href="https://doi.org/10.1021/bi0611762">Link</a> </p> | ||
| + | <p> Wei, Xin-Yuan; Sakr, Samer; Li, Jian-Hong; Wang, Li; Chen, Wen-Li; Zhang, Cheng-Cai (2006): Expression of split dnaE genes and trans-splicing of DnaE intein in the developmental cyanobacterium Anabaena sp. PCC 7120. In: Research in Microbiology 157 (3). <a href="https://doi.org/10.1016/j.resmic.2005.08.004"> Link</a> </p> | ||
| + | <p> Sali, A., & Webb, B. (1989): MODELLER (Version 9.22). Retrieved from https://salilab.org/modeller/ </p> | ||
| + | <p> Ingebrigtsen, Sveinung G.; Didriksen, Alena; Johannessen, Mona; Skalko-Basnet, Natasa; Holsaeter, Ann Mari (2017): Old drug, new wrapping – A possible comeback for chloramphenicol? In: International Journal of Pharmaceutics 526 (1-2). <a href="https://doi.org/10.1016/j.ijpharm.2017.05.025">Link</a> </p> | ||
| + | <p> Shaw, W. V.; Leslie, A. G. W. (1991): Chloramphenicol Acetyltransferase. In: Annual Review of biophysics. <a href="https://www.annualreviews.org/doi/pdf/10.1146/annurev.bb.20.060191.002051">Link</a> </p> | ||
| + | <p> Shaw, William V. (1983): Chloramphenicol Acetyltransferase: Enzymology and Molecular Biology. In: Molecular Biology 14 (1). <a href="https://doi.org/10.3109/10409238309102789">Link</a> </p> | ||
| + | <p> Pongs, O. (1979): Chapter 3: Chloramphenicol". In Hahn, eFred E. (ed.). Mechanism of Action of Antibacterial Agents. In: Antibiotics Volume V Part 1. <a href=" ISBN 978-3-642-46403-4."></a> </p> | ||
| + | <p> Edwards, Keith H. (2009): Optometry: Science, Techniques and Clinical Management. In: Elsevier Health Sciences. <a href=" ISBN 978-0750687782"></a> </p> | ||
| + | <p> Volkmann, Gerrit; Iwai, Hideo (2010): Protein trans-splicing and its use in structural biology: opportunities and limitations. In: Molecular BioSystems 11 <a href="10.1039/C0MB00034E"></a> </p> | ||
| + | <p> Wu, Hong; Hu, Zhuma; Liu, Xiang-Qin (1998): Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. In: Proceedings of the National Academy of Sciences of the United States of America 95 (16) <a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC21320/pdf/pq009226.pdf"></a> </p> | ||
| + | </details> | ||
| + | </div> <!-- End content block --> | ||
| + | </div> | ||
| + | </div> <!-- End Wrapper--> | ||
| + | <div id="page-arrow-top" onclick="topFunction()" style="opacity: 1;"> </div> | ||
| + | |||
| + | <!--Script BackToTop--> | ||
| + | <script> | ||
| + | var mybutton = document.getElementById("page-arrow-top"); | ||
| + | |||
| + | window.onscroll = function() {scrollFunction(),StickyFunction()}; | ||
| + | |||
| + | function scrollFunction() { | ||
| + | if (document.body.scrollTop > 60 || document.documentElement.scrollTop > 60) { | ||
| + | mybutton.style.display = "block"; | ||
| + | } else { | ||
| + | mybutton.style.display = "none"; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | function topFunction() { | ||
| + | document.body.scrollTop = 0; | ||
| + | document.documentElement.scrollTop = 0; | ||
| + | } | ||
| + | </script> | ||
| + | |||
| + | <script> <!--Script for processbar please do not change anything--> | ||
| + | (function(){ | ||
| + | var $w = $(window); | ||
| + | var $circ = $('.animated-circle'); | ||
| + | var $circZwei= $('.animated-circle-zwei'); | ||
| + | var $circDrei= $('.animated-circle-drei'); | ||
| + | var $circVier= $('.animated-circle-vier'); | ||
| + | var $circFünf= $('.animated-circle-fünf'); | ||
| + | var $circSechs= $('.animated-circle-sechs'); | ||
| + | var $circSieben= $('.animated-circle-sieben'); | ||
| + | |||
| + | var wh, h, sHeight, partOne, partTwo, partThree, partFour,partFive, partSix, partSeven; | ||
| + | |||
| + | function setSizes1(){ | ||
| + | wh = $w.height()+150; | ||
| + | s=$("#a1").offset().top+$(window).innerHeight(); //ist der Abstand vom Anfang der Seite bis nach der Summary | ||
| + | s2=$("#a2").offset().top+$(window).innerHeight();//+140; //Erster Abschnitt h1- H2 | ||
| + | s3=$("#a3").offset().top+$(window).innerHeight();//-50; | ||
| + | s4=$("#a4").offset().top+$(window).innerHeight();//-50; | ||
| + | s5=$("#a5").offset().top+$(window).innerHeight();//-50; | ||
| + | s6=$("#a6").offset().top+$(window).innerHeight(); | ||
| + | s7=($("#Ende").offset().top)+250; //zweiter Abschnitt h2-h3 | ||
| + | h = $('body').height()-$(window).innerHeight(); | ||
| + | partOne =s-wh; | ||
| + | partTwo= s2-(wh+partOne);//wh; | ||
| + | partThree=s3-(wh+partTwo+partOne); | ||
| + | partFour=s4-(wh+partThree+partTwo+partOne); | ||
| + | partFive=s5-(wh+partFour+partThree+partTwo+partOne); | ||
| + | partSix=s6-(wh+partFive+partFour+partThree+partTwo+partOne); | ||
| + | partSeven=s7-(wh+partSix+partFive+partFour+partThree+partTwo+partOne); | ||
| + | } | ||
| + | |||
| + | setSizes1(); | ||
| + | |||
| + | $w.on('scroll', function(){ | ||
| + | var percOne = Math.max(0, Math.min(1, $w.scrollTop()/partOne)); | ||
| + | var percTwo = Math.max(0, Math.min(1, ($w.scrollTop()-partOne)/partTwo)); | ||
| + | var percThree = Math.max(0, Math.min(1, ($w.scrollTop()-partTwo-partOne)/partThree)); | ||
| + | var percFour = Math.max(0, Math.min(1, ($w.scrollTop()-partThree-partTwo-partOne)/partFour)); | ||
| + | var percFive = Math.max(0, Math.min(1, ($w.scrollTop()-partFour-partThree-partTwo-partOne)/partFive)); | ||
| + | var percSix = Math.max(0, Math.min(1, ($w.scrollTop()-partFive-partFour-partThree-partTwo-partOne)/partSix)); | ||
| + | var percSeven= Math.max(0, Math.min(1, ($w.scrollTop()-partSix-partFive-partFour-partThree-partTwo-partOne)/partSeven)); | ||
| + | updateProgress1(percTwo,percThree,percFour,percFive,percSix, percSeven); | ||
| + | }).on('resize', function(){ | ||
| + | setSizes1(); | ||
| + | $w.trigger('scroll'); | ||
| + | }); | ||
| + | |||
| + | |||
| + | function updateProgress1(percTwo,percThree,percFour,percFive,percSix,percSeven){ | ||
| + | |||
| + | var circle_offset = 63 * percTwo; | ||
| + | var circle_offset_zwei = 63 * percThree; | ||
| + | var circle_offset_drei = 63 * percFour; | ||
| + | var circle_offset_vier = 63 * percFive; | ||
| + | var circle_offset_fünf = 63 * percSix; | ||
| + | var circle_offset_sechs = 63 * percSeven; | ||
| + | $circZwei.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset | ||
| + | }); | ||
| + | $circDrei.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset_zwei | ||
| + | }); | ||
| + | $circVier.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset_drei | ||
| + | }); | ||
| + | |||
| + | $circFünf.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset_vier | ||
| + | }); | ||
| + | |||
| + | $circSechs.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset_fünf | ||
| + | }); | ||
| + | |||
| + | $circSieben.css({ | ||
| + | "stroke-dashoffset" : 63 - circle_offset_sechs | ||
| + | }); | ||
| + | |||
| + | |||
| + | } | ||
| + | |||
| + | }()); | ||
| + | |||
| + | </script> | ||
| + | |||
| + | |||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 03:40, 22 October 2019
Chloramphenicol resistance protein variant B Part 2
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Chloramphenicol is an antibiotic which easily diffuses across cell membranes.
Once inside the cell it interferes in the protein biosynthesis pathway.
More precisely it binds to specific residues of the 23S rRNA of the 50S ribosomal subunit
and interrupts the formation of new peptide bonds.
In 1947 chloramphenicol was first discovered in an isolate from Streptomyces venezuelae,
later that decade it became the first artificially produced antibiotic. (Pong 1979)
In every day life chloramphenicol serves as a back up broadband antibiotic which is used in case other
antibiotics can not be applied. Normally it is only used in case of need, since it comes with a broad range of side effects.
Nevertheless, chloramphenicol is often one of the ingredients in eye ointments for the treatment of conjunctivitis. (Edwards 2009)
Furthermore it is used to treat hazardous bacterial infections like plaque, cholera, MRSA or typhoid fever. (Ingebrigtsen 2017)
Additionally, it is one of the most commonly used antibiotics in molecular biology for the selection of genetically modified organisms.
Many of the vectors for gene transfer used in research carry a gene encoding a chloramphenicol resistance.
Thus, if a cell has taken up the vector which is encoding the gene of interest as well as the resistance gene this cell
becomes resistant against chloramphenicol.
Resistance
Intein-mediated protein splicing.
Inteins
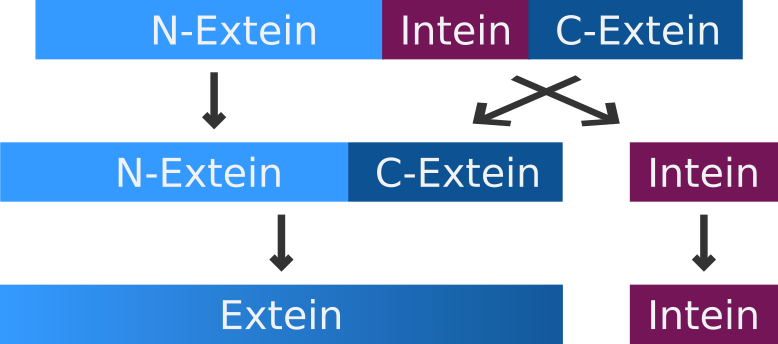
The intein-mediated protein splicing occurs after the mRNA is translated into a sequence of amino acids. Prior to the splicing process the N-terminal part of the precursor protein is called N-extein, the center part is the intein and the C-terminal part is named C-extein. The spliced protein is called an extein as well.
Split inteins
A very special but small subset of inteins are the so-called split-inteins. They are transcribed and translated as separate polypeptides but rapidly associate afterwards to form the active intein. The active intein then ligates the fused N- and C-exteins in a process called protein trans-splicing.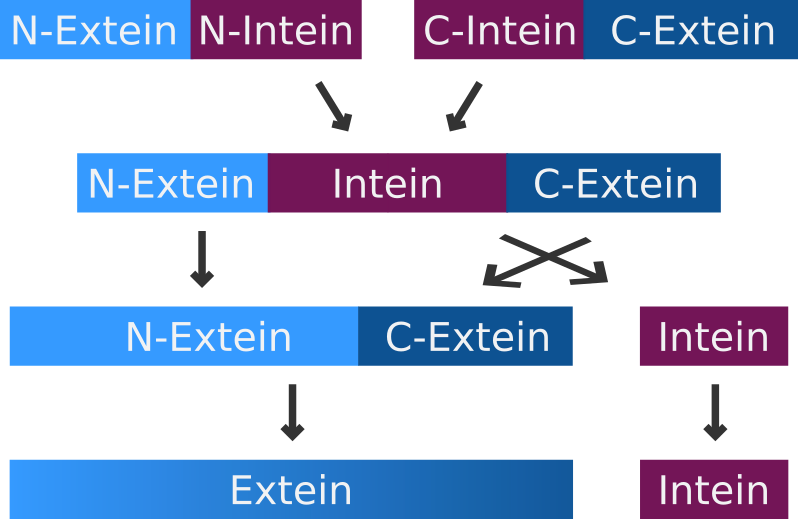
Amino acids neighboring split sites


Mechanism
For a better understanding of the importance of the amino acid residues near the splicing site, following is a short summary of the split-intein-mediated splicing mechanism::
- Split inteins associate
- Nucleophilic attack of the peptide bond between the N-extein and the intein by the residue of the first amino acid in the intein
- Formation of a linear ester (not shown) or thioester intermediate
- Side chain of the first residue of the C-extein attacks the thioester (or ester), freeing the N-terminal end of the intein
- N- and C-extein are connected, but not yet by a peptide bond
- Amide nitrogen atom from the residue of the last amino acid of the intein cleaves the peptide bond between the intein and the C-extein
- Intein is free
- Free amino group of the C-extein attacks thioester (or ester) linking the exteins together
- O-N or S-N shift forms a peptide bond creating a fully functional protein
Split-resistance genes
Split inteins enable us to divide and fuse all kinds of proteins. Their application ranges from structural biology, especially NMR spectroscopy to split intein-mediated production of genetically encoded cyclic peptide libraries. (Horshill and Benkovic 2006; Volkmann and Iwai 2010) Another possible application are split markers, like antibiotics for the selection of GMOs.
Researchers working in the field of synthetic biology are often using two or multiple plasmid systems to incorporate all desired transgenes into one cell. For the selection of positive clones and stable retention of the plasmids they usually apply several antibiotics to the cultivation medium. Unfortunately, multiple antibiotics provide very harsh growth conditions for the cells.
Split antibiotics are a great solution to reduce the number of needed antibiotics. A scheme how split markers are implemented is illustrated in figure 8. To create a split selectable marker the gene encoding an antibiotic resistance is split into two segments. Each segment is fused to one part of a split intein which is able to join the protein segments back together. Each of these fusion segments is inserted into a different vector. If a cell is transformed with both cohesive vectors it is able to develop a resistance against the respective antibiotic via protein trans-splicing. If the cell has taken up only one of the vectors it has no chance to survive selection since it can produce only one part of the protein mediating the antibiotic resistance.

Biosafety
When using split selectable markers one part of the protein is encoded by a plasmid whereas the other part could be encoded by either another plasmid or the genome of the target organism. In both cases this system would increase the biosafety of the systems encoded on the plasmids. If one of the plasmids, a strain containing only one of the plasmids, or a strain encoding one of the parts in its genome would be released into the environment the antibiotic resistance would not spread. Since both parts of the protein are needed to establish the resistance, the uptake of only one of the parts would not increase the fitness of an organism in the outer world. This reduces the probability of GMOs to overgrow natural populations in the environment. Therefore, this system could increase the safety of GMOs in any application outside of the lab. This would be beneficial for the future application of many iGEM projects.
Model and plasmid design
Preliminary considerations
To assemble our Troygenics we are working with a two plasmid system. Since most of the real world applications of the Troygenics involve their release into the outer world, the biosafety of our system is a crucial part of our project. As explained above split selectable markers are a great way to improve the biosafety of two plasmid systems, because they not only reduce the likelihood of spread antibiotic resistances but also reduce the probability of GMOs overgrowing naturally occurring populations. As chloramphenicol is the standard antibiotic resistance used by the iGEM community but was never submitted as a split antibiotic resistance before we decided to develop a split chloramphenicol resistance, which is particular useful for future iGEM teams. It was our aim to split the resistance gene cat so that the two parts of the chloramphenicol acetytransferase are encoded by two different plasmids. According to the advantages mentioned above, we decided to use the split intein NpuDnaE (Partnummern)to create our kanamycin split resistance.The important amino acids neighboring the native splice site of NpuDnaE are“Ala Glut Tyr” for the N-extein and “Cys Phe Asn” for the native C-extein. (Cheriyan et al. 2013) To examine the optimal split point of the chloramphenicol acetyltransferase which mediates the chloramphenicol resistance we designed and executed a model. It provides us with a list of all possible split points, ranked from best to worst, for the desired protein, based on its amino acid sequence and 3D structure.
Modeling the optimal split point
| Amino Acid | rF(Ai) at the N-terminal extein | rF(Ai) at the C-terminal extein | ||||
|---|---|---|---|---|---|---|
| -3 | -2 | -1 | +1 | +2 | +3 | |
| D | 1.39 | 1.52 | 0.00 | 0.00 | 0.00 | 1.26 |
| E | 1.26 | 3.51 | 0.07 | 0.00 | 0.00 | 5.5 |
| N | 0.93 | 1.26 | 2.19 | 0.00 | 0.00 | 8.61 |
| Q | 1.39 | 0.86 | 0.33 | 0.00 | 0.00 | 0.27 |
| H | 1.06 | 1.26 | 2.19 | 0.00 | 0.00 | 8.61 |
| K | 1.59 | 0.93 | 4.44 | 0.00 | 0.07 | 0.00 |
| R | 1.81 | 0.2 | 0.86 | 0.00 | 0.00 | 0.00 |
| S | 0.99 | 1.24 | 1.37 | 0.00 | 0.00 | 0.07 |
| C | 0.46 | 0.40 | 0.80 | 50 | 0.07 | 0.07 |
| T | 0.63 | 0.89 | 1.59 | 0.00 | 0.00 | 0.96 |
| P | 0.80 | 1.52 | 0.00 | 0.00 | 0.00 | 0.00 |
| G | 4.67 | 2.68 | 0.17 | 0.00 | 0.00 | 0.03 |
| A | 0.63 | 0.83 | 1.92 | 0.00 | 0.00 | 0.00 |
| V | 0.56 | 0.50 | 0.23 | 0.00 | 0.03 | 0.10 |
| I | 0.07 | 0.53 | 0.00 | 0.00 | 0.00 | 0.40 |
| L | 0.11 | 0.57 | 0.75 | 0.00 | 0.00 | 0.82 |
| M | 0.13 | 0.73 | 1.26 | 20 | 0.07 | 2.25 |
| F | 0.13 | 0.73 | 1.26 | 0.00 | 0.07 | 2.25 |
| Y | 0.20 | 0.60 | 2.05 | 0.00 | 0.13 | 5.23 |
| W | 0.07 | 0.40 | 0.99 | 0.00 | 29.42 | 0.07 |

Using this approach, a list with all possible split points sorted by B(seq)n is created, ranging from the best to the worst possible split point in the protein.
Complementing the sequence based determination of potential split points the desired structure of the final protein has to be considered. Positions in important structural features of the protein were determined using the protein structure viewer Chimera (Pettersen et al., 2004). Sequences involved in relevant structures were integrated in the analyses as described above to prevent the identification of possible split points in these regions. Lastly, the remaining list of split points was sorted from best to worst and the best split sequences determined were written FASTA files. Using the MODELLER-software (Sali & Webb, 1989), the most likely structure of the N- or C-terminal part of the protein fused to the split intein, prior to finding the other intein in the cell, was determined. To do so, it was necessary to find proteins with similar structures as templates for homology modeling. They were chosen as follows:
Split Chloramphenicol Resistance
Even though this split point is located at the second amino acid of a beta-sheet, we decided to test whether splitting at this position could lead to Chloramphenicol acetyltransferase fragments that could be reconstituted by intein-mediated protein splicing.
Similar to our modeling of the split kanamycin resistance, we used the homology modeling software MODELLER (Sali & Webb, 1989) to visualize each part of the chloramphenicol acetyltransferase fused to the Npu DnaE intein.
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCLSYETEILTVEYGLLPIGKIVEKRIECTVYSVDNNGNIYTQPVAQWHDRGEQEVFEYCLEDGSLIRATKDHKFMTVDGQMLPIDEIFERELDLMRVDNLPNNLEGHHHHHH
Part 2:
MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNCCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
The templates used for homology modeling were:
| Part 1 | Part 2 |
|---|---|
| 1NOC, chain B (Chloramphenicol acetyltransferase) | 1NOC, chain B (Chloramphenicol acetyltransferase) |
| 4QFQ, chain A (Npu DnaE) | 4QFQ, chain B (Npu DnaE) |
| 5OL6, chain A (inactivated Npu SICLOPPS intein with CAFHPQ extein) | 5OL6, chain B (inactivated Npu SICLOPPS intein with CAFHPQ extein) |
| 4KL5, chain A (Npu DnaE) | 1QCA, chain A (Type III Chloramphenicol acetyltransferase) |
| 2KEQ, chain A (DnaE intein from Nostoc punctiforme) | 3CLA, chain A (Type III Chloramphenicosl acetyltransferase) |
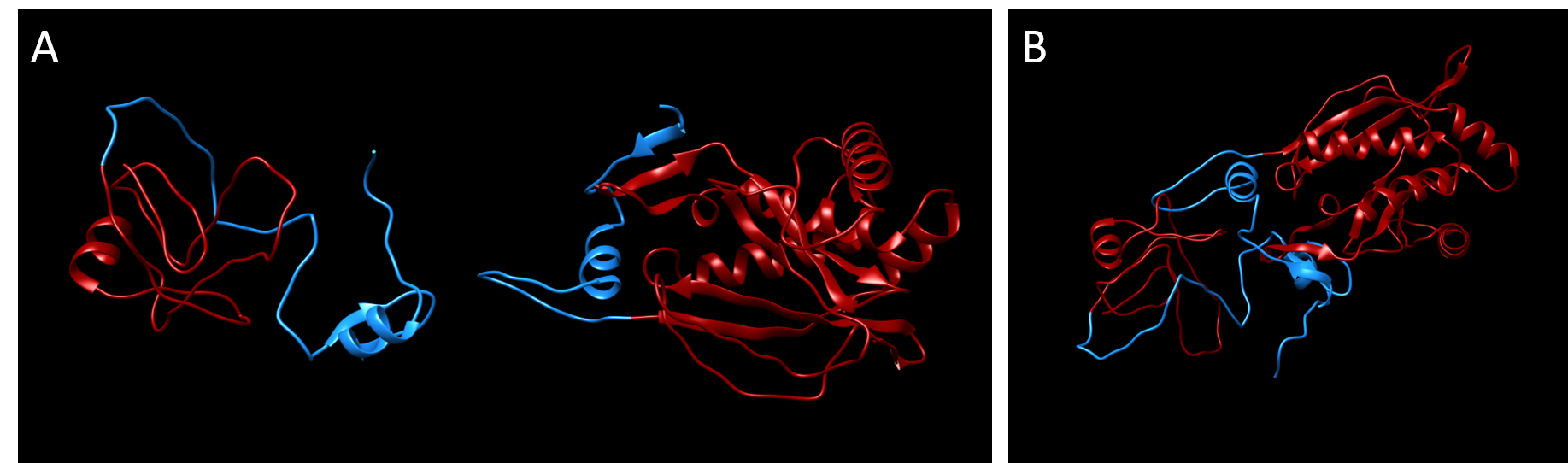
Plasmid design
We designed 4 parts encoding this chloramphenicol split resistance. Part BBa_2926076 and BBa_2926076 are basic parts. BBa_2926076 is encoding the N-terminal part of the split resistance, fused to the N-intein. This part was constructed in pSB3T5. Whereas BBa_2926076 encodes the C-intein fused to the C-terminal part of the split resistance and was constructed in pSB1K3.Additionally to the split chloramphenicol resistance described on this page, we have constructed another chloramphenicol split resistance, as well as a kanamycin, ampicilin and a hygromycin split resistance. Here you can get more information on our handy collection of split antibiotic resistances.
Characterization
We have successfully constructed all parts and verified their nucleotide sequence with sanger sequencing.
After co-transformation of BBa_2926076 and BBa_2926076 into E. coli DH5α the cells where plated on selection plates containing kanamycin and tetracyclin, but no chloramphenicol since the cells need some time to establish the chloramphenicol resistance.
Afterwards a number of colonies were transferred to numerous plates, each containing a different concentration from 0.1 up to 100 μg/mL chloramphenicol. Alongside with our colonies containing the split chloramphenicol resistance we plated E. coli DH5α as a negativ control and E. coli DH5α containing pSB1C3, which is encoding RFP as a positive control.
We conducted this experiment as a triplicate. The results of one of the triplicates are depicted in figure 10. Native E. coli DH5α was only growing on the plates containing 0.1 μg/mL chloramphenicol, implying that the native E. coli DH5α has no significant resistance to chloramphenicol. E. coli DHα containing pSB1C3, which is encoding RFP was proven to be resistant on all tested chloramphenicol concentrations. The E. coli DH5α encoding our split chloramphenicol resistance showed to be resistant to chloramphenicol concentrations of averaged 7 μg/mL.
In every day lab life chloramphenicol is used in concentrations between 10 and 25 μg/mL. Since our split system shows resistance against chloramphenicol for concentrations of up to averaged 7 μg/mL the working concentrations for chloramphenicol could be reduced, leading to a diminished chloramphenicol consumption.
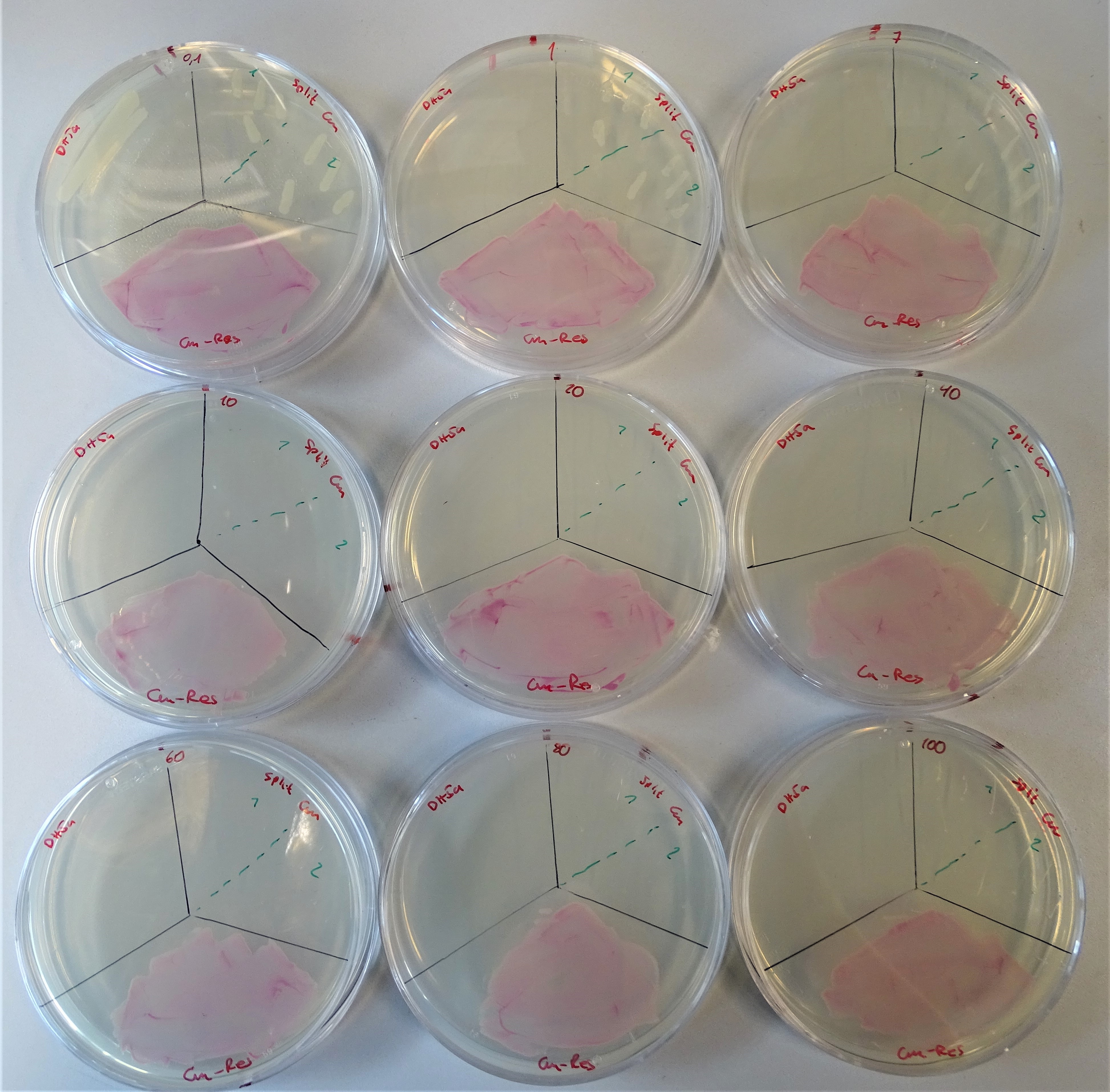
List of our basic Parts
| Basic parts | Description | Designer | Length |
|---|---|---|---|
| BBa_K2926000 | Cas13a Lsh | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 4168 bp |
| BBa_K2926001 | Cas13a Lbu | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 3480 bp |
| BBa_K2926003 | GALL yeast promoter (validated part) | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 430 bp |
| BBa_K2926004 | SNR52 yeast promoter | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 269 bp |
| BBa_K2926005 | TPS1 yeast terminator | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 250 bp |
| BBa_K2926006 | SUP4 yeast terminator | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 20 bp |
| BBa_K2926007 | Inverted terminal repeat form S. cerevisiae gen ADE2, front | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 50 bp |
| BBa_K2926008 | gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA1-3 | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 187 bp |
| BBa_K2926009 | gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA1-7 | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 435 bp |
| BBa_K2926010 | gRNA array (cas13a) with sgRNAs (for S.cerevisiae) gRNA4-7 | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 301 bp |
| BBa_K2926011 | gRNA array (cas13a) with 3 sgRNAs (for A. niger) | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 207 bp |
| BBa_K2926013 | Inverted terminal repeat form S. cerevisiae gen ADE2, back | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 50 bp |
| BBa_K2926015 | Aspergillus niger 5S rRNA | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 119 bp |
| BBa_K2926020 | Promotor of protein VIII from the M13 phage | Astrid Többer | 47 bp |
| BBa_K2926021 | sgRNA for the detection of RFP with Cas13a | Ina Schmitt, Isabel Conze, Katharina Wolff, Johanna Opgenoorth | 57 bp |
| BBa_K2926022 | Trp_sfGFP_Opy2p_mCherry_M13K07 gene VIII_M13K07 gene III | Johanna Opgenoorth | 4066 bp |
| BBa_K2926023 | M13K07_genes_II_VIII | Astrid Többer | 2015 bp |
| BBa_K2926025 | M13K07_genes_III_IV | Astrid Többer | 2820 bp |
| BBa_K2926026 | M13K07_truncated_III | Astrid Többer | 456 bp |
| BBa_K2926027 | mCherry_VIII_fusion_protein | Astrid Többer | 1086 bp |
| BBa_K2926048 | mCherry with hexahistidine tag for purification | Johanna Opgenoorth | 729 bp |
| BBa_K2926049 | Mating factor alpha from S. cerevisiae fused to mCherry | Johanna Opgenoorth | 762 bp |
| BBa_K2926050 | Fusion protein of Flo11 from yeast and Cherry | Johanna Opgenoorth | 1287 bp |
| BBa_K2926051 | Fusion protein of Opy2 from yeast and Cherry | Johanna Opgenoorth | 837 bp |
| BBa_K2926052 | Fusion protein of mCherry and M13 bacteriophage protein pVIII | Johanna Opgenoorth | 942 bp |
| BBa_K2926053 | Fluorescence reporter mCherry fused to pVIII from M13 bacteriophage with purification tag | Johanna Opgenoorth | 729 bp |
| BBa_K2926054 | Fusion protein of mating factor alpha from yeast, mCherry, and pVIII from M13 bacteriophage | Johanna Opgenoorth | 994 bp |
| BBa_K2926055 | Fusion protein of Flo11 from yeast, mCherry, and pVIII from M13 bacteriophage | Johanna Opgenoorth | 1521 bp |
| BBa_K2926056 | Fusion protein of Opy2 from yeast, mCherry, and pVIII from M13 bacteriophage | Johanna Opgenoorth | 1071 bp |
| BBa_K2926057 | Fusion protein of mating factor alpha, mCherry, and pVIII with His-tag | Johanna Opgenoorth | 1014 bp |
| BBa_K2926058 | Fusion protein of Flo11 (yeast), mCherry, and pVIII (M13 bacteriophage) with purification tag | Johanna Opgenoorth | 1539 bp |
| BBa_K2926067 | Fusion protein of Opy2 (yeast), mCherry and pVIII (bacteriophage M13) with purification tag | Johanna Opgenoorth | 1089 bp |
| BBa_K2926068 | Proline-glycine-peptide fused to the n-terminus of mCherry | Johanna Opgenoorth | 750 bp |
| BBa_K2926076 | Beta-Cmp resistance protein part 1 (Cm_B_split1_NpuN_pSB3Cd5_Kan)/td> | Nefeli Chanoutsi | 537 bp |
| BBa_K2926077 | Beta-Cmp resistance protein part 2 (NpuC_Cm_B_split2_pSB1Cc3_Tet)/td> | Nefeli Chanoutsi | 543 bp |
| BBa_K2926078 | Alpha-Kan resistance protein part 1 (Kan_A_split1_NpuN_pSB3Kb5_Cm) | Nefeli Chanoutsi | 591 bp |
| BBa_K2926079 | Alpha-Kan resistance protein part 2 (NpuC_Kan_A_split2_pSB1Ka3_Tet) | Nefeli Chanoutsi | 645 bp |
| BBa_K2926080 | Alpha-Amp resistance protein part 1 (Amp_A_Split1_NpuN_pSB1Aa3_Tet) | Nefeli Chanoutsi | 645 bp |
| BBa_K2926081 | Alpha-Amp resistance protein part 2 (NpuC_Amp_A_split2_pSB3Ab5_Cm) | Nefeli Chanoutsi | 645 bp |
References
Shah, Neel H.; Muir, Tom W. (2011): Split Inteins: Nature's Protein Ligases. In: Israel Journal of Chemistry 51 (8-9). Link
Zettler, Joachim; Schütz, Vivien; Mootz, Henning D. (2009): The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. In: FEBS Letters 583. Link
Jillette, Nathaniel; Du, Menghan; Cheng, Albert (2019): Split Selectable Markers. In: bioRxiv. Link
Iwai, Hideo; Züger, Sara; Jin, Jennifer; Tam, Pui-Hang (2006): Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. In: FEBS Letters 580 (7). Link
Cheriyan, Manoj; Pedamallu, Chandra S.; Tori, Kazuo; Perler, Francine (2013): Faster Protein Splicing with the Nostoc punctiforme DnaE Intein Using Non-native Extein Residues. In: Journal of Biological Chemistry 288 (6). Link
Oeemig, Jesper S.; Aranko, A. Sesilja; Djupsjöbacka, Janica; Heinämäki, Kimmo; Iwai, Hido (2009): Solution structure of DnaE intein from Nostoc punctiforme: Structural basis for the design of a new split intein suitable for site‐specific chemical modification. In: FEBS Letters 583 (9). Link
Dassa, Bareket; Amitai, Gil; Caspi, Jonathan; Schueler-Furman, Ora; Pietrokovski, Shmuel (2007): Trans Protein Splicing of Cyanobacterial Split Inteins in Endogenous and Exogenous Combinations. In: Biochemistry 46 (1). Link
Wei, Xin-Yuan; Sakr, Samer; Li, Jian-Hong; Wang, Li; Chen, Wen-Li; Zhang, Cheng-Cai (2006): Expression of split dnaE genes and trans-splicing of DnaE intein in the developmental cyanobacterium Anabaena sp. PCC 7120. In: Research in Microbiology 157 (3). Link
Sali, A., & Webb, B. (1989): MODELLER (Version 9.22). Retrieved from https://salilab.org/modeller/
Ingebrigtsen, Sveinung G.; Didriksen, Alena; Johannessen, Mona; Skalko-Basnet, Natasa; Holsaeter, Ann Mari (2017): Old drug, new wrapping – A possible comeback for chloramphenicol? In: International Journal of Pharmaceutics 526 (1-2). Link
Shaw, W. V.; Leslie, A. G. W. (1991): Chloramphenicol Acetyltransferase. In: Annual Review of biophysics. Link
Shaw, William V. (1983): Chloramphenicol Acetyltransferase: Enzymology and Molecular Biology. In: Molecular Biology 14 (1). Link
Pongs, O. (1979): Chapter 3: Chloramphenicol". In Hahn, eFred E. (ed.). Mechanism of Action of Antibacterial Agents. In: Antibiotics Volume V Part 1.
Edwards, Keith H. (2009): Optometry: Science, Techniques and Clinical Management. In: Elsevier Health Sciences.
Volkmann, Gerrit; Iwai, Hideo (2010): Protein trans-splicing and its use in structural biology: opportunities and limitations. In: Molecular BioSystems 11
Wu, Hong; Hu, Zhuma; Liu, Xiang-Qin (1998): Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. In: Proceedings of the National Academy of Sciences of the United States of America 95 (16)
