Difference between revisions of "Part:BBa K3254025"
(→Thermodynamic Characterization) |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3254025 short</partinfo> | <partinfo>BBa_K3254025 short</partinfo> | ||
| − | This sfGFP generator could respond to IPTG concentration. This device would have an appropriate output range on low copy plasmid. This is also an example of the composite application of lacI expression cassette([[Part:BBa_K3254022|BBa_K3254022]]), Ptac promoter([[Part:BBa_K3254014|BBa_K3254014]]) and RiboJ([[Part:BBa_K2615996|BBa_K2615996]]). | + | *This sfGFP generator could respond to IPTG concentration. This device would have an appropriate output range on low copy plasmid. This is also an example of the composite application of lacI expression cassette([[Part:BBa_K3254022|BBa_K3254022]]), Ptac promoter([[Part:BBa_K3254014|BBa_K3254014]]/[[Part:BBa_K2572025|BBa_K2572025]]) and RiboJ([[Part:BBa_K2615996|BBa_K2615996]]). |
| + | *The response curve of this operon can be used as a standard reference of the Ptac promoter's output. | ||
| + | *Other translational units can replace the [[Part:BBa_K3254024|BBa_K3254024]] for constructing useful inducible expression systems. | ||
| + | |||
| + | =Thermodynamic Characterization= | ||
| + | *'''Group:''' GENAS_China 2019 | ||
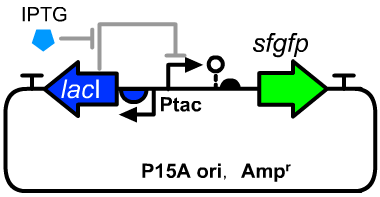
| + | *We constructed this operon on a P15A plamisd. | ||
| + | *A series of IPTG inducible promoters with different dynamic ranges and non-induced activities could be achieved just by replacing the [[Part:BBa_K3254014|BBa_K3254014]](twin: [[Part:BBa_K2572025|BBa_K2572025]]) with different Ptac derived promoters, including [[Part:BBa_K864400|Original Ptac]], [[Part:BBa_K3254015|Ptac-M1]], [[Part:BBa_K3254016|Ptac-M2]] and [[Part:BBa_K3254017|Ptac-M3]]. | ||
| + | |||
| + | ==Genetic Design== | ||
| + | [[File:T--GENAS_China--Ptac-sfGFP_plasmid.PNG|200px|thumb|left|The structure of the experimental plasmid]] | ||
| + | *The sequence of [[Part:BBa_K3254025|BBa_K3254025]] can be seen as a reference. | ||
| + | *The host cell was E.coli DH5α. | ||
| + | |||
| + | ==Experimental Setup== | ||
| + | *All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using Corning flat-bottom 96-well plates sealed with sealing film. For characterization the circuit response functions, a previously developed quantitative method that measured gene expression at steady state was used(Zhang, Chen et al. 2016). Briefly, bacteria harboring the parts/circuits of interest were first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth, after which the cell cultures were diluted 196-fold with M9 medium. After 3 h of growth, the cultures were further diluted 700-fold with M9 medium containing gradient concentrations of IPTG, and incubated for another 6 h. Finally, 20-μL samples of each culture were transferred to a new plate containing 180 μL per well of PBS supplemented with 2 mg/mL kanamycin to terminate protein expression. The fluorescence distribution of each sample was assayed using a flow cytometer with appropriate voltage settings; each distribution contained >20,000 events. Each sample was experimentally assayed at least three times. The arithmetical mean of each sample was determined using FlowJo software. | ||
| + | *M9 medium (supplemented): 6.8 g/L Na<sub>2</sub>HPO<sub>4</sub>, 3 g/L KH<sub>2</sub>PO<sub>4</sub>, 0.5 g/L NaCl, 1 g/L NH<sub>4</sub>Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO<sub>4</sub>, and 100 μM CaCl<sub>2</sub>. | ||
| + | |||
| + | ==Results== | ||
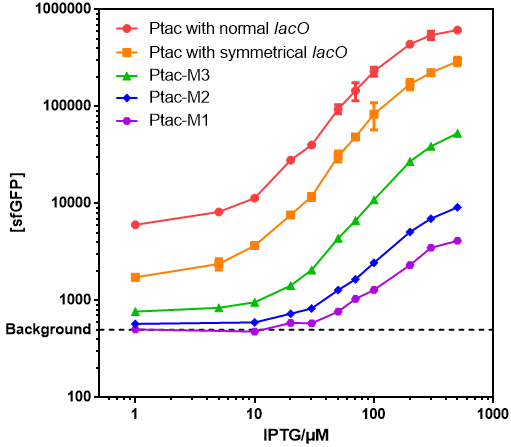
| + | *Those promoters had gentle response curves without saltus. | ||
| + | [[File:T--GENAS_China--IPTG-sfGFP.PNG]] | ||
| + | *Background: Cells without any FP genes. | ||
| + | *Ptac with normal lacO: [[Part:BBa_K864400|BBa_K864400]] | ||
| + | *Ptac with symmetrical lacO: [[Part:BBa_K3254014|BBa_K3254014]](twin: [[Part:BBa_K2572025|BBa_K2572025]]) | ||
| + | *Ptac-M1: [[Part:BBa_K3254015|BBa_K3254015]] | ||
| + | *Ptac-M2: [[Part:BBa_K3254016|BBa_K3254016]] | ||
| + | *Ptac-M3: [[Part:BBa_K3254017|BBa_K3254017]] | ||
| + | |||
| + | ==References== | ||
| + | *Zhang HM, et al. Measurements of Gene Expression at Steady State Improve the Predictability of Part Assembly. ACS Synthetic Biology 5, 269-273 (2016). | ||
| + | *Zong Y, et al. Insulated transcriptional elements enable precise design of genetic circuits. Nature communications 8, 52 (2017). | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 11:31, 21 October 2019
Ptac-RiboJ-RBS-sfGFP-teminator
- This sfGFP generator could respond to IPTG concentration. This device would have an appropriate output range on low copy plasmid. This is also an example of the composite application of lacI expression cassette(BBa_K3254022), Ptac promoter(BBa_K3254014/BBa_K2572025) and RiboJ(BBa_K2615996).
- The response curve of this operon can be used as a standard reference of the Ptac promoter's output.
- Other translational units can replace the BBa_K3254024 for constructing useful inducible expression systems.
Thermodynamic Characterization
- Group: GENAS_China 2019
- We constructed this operon on a P15A plamisd.
- A series of IPTG inducible promoters with different dynamic ranges and non-induced activities could be achieved just by replacing the BBa_K3254014(twin: BBa_K2572025) with different Ptac derived promoters, including Original Ptac, Ptac-M1, Ptac-M2 and Ptac-M3.
Genetic Design
- The sequence of BBa_K3254025 can be seen as a reference.
- The host cell was E.coli DH5α.
Experimental Setup
- All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using Corning flat-bottom 96-well plates sealed with sealing film. For characterization the circuit response functions, a previously developed quantitative method that measured gene expression at steady state was used(Zhang, Chen et al. 2016). Briefly, bacteria harboring the parts/circuits of interest were first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth, after which the cell cultures were diluted 196-fold with M9 medium. After 3 h of growth, the cultures were further diluted 700-fold with M9 medium containing gradient concentrations of IPTG, and incubated for another 6 h. Finally, 20-μL samples of each culture were transferred to a new plate containing 180 μL per well of PBS supplemented with 2 mg/mL kanamycin to terminate protein expression. The fluorescence distribution of each sample was assayed using a flow cytometer with appropriate voltage settings; each distribution contained >20,000 events. Each sample was experimentally assayed at least three times. The arithmetical mean of each sample was determined using FlowJo software.
- M9 medium (supplemented): 6.8 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO4, and 100 μM CaCl2.
Results
- Those promoters had gentle response curves without saltus.
- Background: Cells without any FP genes.
- Ptac with normal lacO: BBa_K864400
- Ptac with symmetrical lacO: BBa_K3254014(twin: BBa_K2572025)
- Ptac-M1: BBa_K3254015
- Ptac-M2: BBa_K3254016
- Ptac-M3: BBa_K3254017
References
- Zhang HM, et al. Measurements of Gene Expression at Steady State Improve the Predictability of Part Assembly. ACS Synthetic Biology 5, 269-273 (2016).
- Zong Y, et al. Insulated transcriptional elements enable precise design of genetic circuits. Nature communications 8, 52 (2017).
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1385