Difference between revisions of "Part:BBa K2455003"
(→Insertion Requirements) |
(→Imaging) |
||
| (81 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2455003 short</partinfo> | <partinfo>BBa_K2455003 short</partinfo> | ||
| − | This biobrick contains an AsiSI/Nb.BSMI USER Cassette with a N-terminal nona-arginine (R9) Cell-penetrating peptide | + | This biobrick contains an AsiSI/Nb.BSMI USER Cassette with a N-terminal nona-arginine (R9) Cell-penetrating peptide. The nona-arginine (R9) gives any cassette inserted proteins the ability to pass through plasma membranes. In addition, the USER Cassette carries a N-terminal hexa-histidine tag which allows for easy purification. |
| − | For our study we demonstrated that proteins inserted into the cassette can be purified | + | For our study we demonstrated that proteins inserted into the cassette can be purified using the histidine-tag, |
| − | and subsequently | + | and subsequently used to stain <i>E. coli</i> and <i>Pseudomonas putida</i> cells, presumably through internalization caused by the R9 peptide. |
| − | In addition; previous studies have shown | + | In addition; previous studies have shown that R9 associated proteins, either covalently or non-covalently, were able to enter a variety of cell types ([https://academic.oup.com/pcp/article/46/3/482/1817347 Chang <i>et al., 2005</i>]). |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 24: | Line 24: | ||
<span class='h3bb'><font size="5"><b>Usage and Biology</b></font></span> | <span class='h3bb'><font size="5"><b>Usage and Biology</b></font></span> | ||
| − | Cell-Penetrating Peptides (CPPs) are small peptides, | + | Cell-Penetrating Peptides (CPPs) are small peptides, that are able to facilitate transport of a wide variety of cargoes across plasma membranes. CPPs are typically rich in basic residues and originate from viral domains such as the viral HIV tat domain ([https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2634491/ Eudes & Chugh, 2008]). In recent years researchers have tried to make synthetic CPPs, with special interest in peptides constructed solely from arginine residues. The arginine rich sequences have been shown to trigger endocytosis in a wide range of cell types, including onion and potato cells ([https://academic.oup.com/pcp/article/46/3/482/1817347 Chang <i>et al.</i>, 2005]). |
==USER Cassette== | ==USER Cassette== | ||
| Line 34: | Line 34: | ||
For more information on USER cloning see [http://www.cbs.dtu.dk/services/AMUSER/ Amuser] or [http://www.cbs.dtu.dk/services/AMUSER/instructions.php Amuser instructions]. | For more information on USER cloning see [http://www.cbs.dtu.dk/services/AMUSER/ Amuser] or [http://www.cbs.dtu.dk/services/AMUSER/instructions.php Amuser instructions]. | ||
| + | |||
| + | ===Insertion Requirements=== | ||
| + | |||
| + | For insertion of genes into the USER Cassette genes have to be amplified with the primers carrying the two following overhangs: | ||
| + | |||
| + | Forward primer: CGTGCGAUCx | ||
| + | |||
| + | Reverse primer: CACGCGAUxx | ||
| + | |||
| + | Where U = Uracil and x = any non-uracil-nucleotide; these are needed to keep the inserted gene in-frame. [https://www.thermofisher.com/order/catalog/product/ER2091?SID=srch-srp-ER2091 AsiSI] and [https://www.neb.com/products/r0706-nbbsmi#Product%20Information Nb.BsmI] have to be used to facilitate opening of the cassette. | ||
==Gene Insertion== | ==Gene Insertion== | ||
| − | In order to test the ability | + | In order to test the biobricks ability to facilitate protein import in <i>E. coli</i> we chose to insert two different fluorescent proteins into the biobrick, [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K592100|mTag BFP]]. |
===Opening of USER Cassette=== | ===Opening of USER Cassette=== | ||
| − | To facilitate insertion we first had to prepare the vector, this was done in a two step digestion reaction. First to open the cassette 42 µL purified vector was mixed with 2.5 µL [https://www.thermofisher.com/order/catalog/product/ER2091?SID=srch-srp-ER2091 AsiSI], 10 µL [https://www.thermofisher.com/order/catalog/product/BY5 10x Tango Buffer], and 45.5 µL Nuclease-free water. The mixture was then incubated for 3 hours at 37 degrees Celsius. 5 µL linearised vector | + | To facilitate insertion, we first had to prepare the vector, this was done in a two-step digestion reaction. First to open the cassette 42 µL purified vector was mixed with 2.5 µL [https://www.thermofisher.com/order/catalog/product/ER2091?SID=srch-srp-ER2091 AsiSI], 10 µL [https://www.thermofisher.com/order/catalog/product/BY5 10x Tango Buffer], and 45.5 µL Nuclease-free water. The mixture was then incubated for 3 hours at 37 degrees Celsius. Then we ran 5 µL of the linearised vector for 20 min at 100 V on a 1 % Agarose gel to check for proper linearization. As shown on Figure 2 the vector goes from a smaller band, typical for circular (supercoiled) pieces of DNA, while the digested vector has a band consistent with the full 6kbp of the vector. |
| − | [[image:PET102_CPP_linearization.JPG|thumb|left|150px|'''Figure 2: | + | [[image:PET102_CPP_linearization.JPG|thumb|left|150px|'''Figure 2: Linearisation of modified pET102 vector carrying the Cell-Penetrating USER Cassette biobrick.''' Lane 1: undigested vector, Lane 2: AsiSI digested vector. Vector has a size of 6kbp.]] |
| − | Following digestion the vector was subjected to column-purified using [http://omegabiotek.com/store/product/e-z-n-a-gel-extraction-kit/ E.Z.N.A.® Gel Extraction Kit] and [http://omegabiotek.com/store/wp-content/uploads/2013/05/D2500.D2501-Gel-Extraction-Kit-Combo-Online.pdf standard protocol] for gel-extraction with 150 | + | Following digestion the vector was subjected to column-purified using [http://omegabiotek.com/store/product/e-z-n-a-gel-extraction-kit/ E.Z.N.A.® Gel Extraction Kit] and [http://omegabiotek.com/store/wp-content/uploads/2013/05/D2500.D2501-Gel-Extraction-Kit-Combo-Online.pdf standard protocol] for gel-extraction with 150 µL binding buffer being added in the first step. |
| − | Finally the vector was nicked by mixing the complete 50 | + | Finally the vector was nicked by mixing the complete 50 µL elute from the column-purification with 1 µL [https://www.neb.com/products/r0706-nbbsmi#Product%20Information Nb.BsmI], 6 µL [https://www.neb.com/products/b7203-nebuffer-3-1#Product%20Information 10x NEBuffer 3.1], and 3 µL Nuclease-free water. The mixture was incubated for 3 hours at 65 degrees Celcius. |
===Gene Amplification=== | ===Gene Amplification=== | ||
| − | In order to facilitate insertion of the [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K592100|mTag BFP]] biobricks into the USER Cassette, the two genes were amplified using the following sets of primers: | + | In order to facilitate the insertion of the [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K592100|mTag BFP]] biobricks into the USER Cassette, the two genes were amplified using the following sets of primers in order to add the appropriate overhangs: |
| + | |||
<b>SYFP2:</b> | <b>SYFP2:</b> | ||
| Line 61: | Line 72: | ||
Reverse primer: CACGCGAUGA-ATTCAGTTTATGACCCAGC | Reverse primer: CACGCGAUGA-ATTCAGTTTATGACCCAGC | ||
| + | |||
| + | And gel-extracted using [http://omegabiotek.com/store/product/e-z-n-a-gel-extraction-kit/ E.Z.N.A.® Gel Extraction Kit] and [http://omegabiotek.com/store/wp-content/uploads/2013/05/D2500.D2501-Gel-Extraction-Kit-Combo-Online.pdf standard protocol]. | ||
| + | |||
| + | |||
===Insertion of BFP and YFP=== | ===Insertion of BFP and YFP=== | ||
| − | + | To insert the gel-extracted PCR products 1 µL of the PCR product was mixed with 2 µL linearised vector, 0.5 µL [https://www.neb.com/products/b7204-cutsmart-buffer#Product%20Information CutSmart® Buffer], 0.5 µL [https://www.neb.com/products/m5505-user-enzyme#Product%20Information USER® Enzyme], and 1 µL MQ H<sub>2</sub>O. The mixture was incubated in a PCR machine for 25 min at 37 degrees Celcius, 20 min at 25 degrees Celcius, 10 min at 20 degrees Celcius, and 10 min at 15 degrees Celcius. | |
| − | == | + | The insertions were validated through PCR amplification of the inserts followed by sequencing. |
| − | === | + | |
| + | ==Purification and SDS-PAGE== | ||
| + | Following the validated insertion of the [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K592100|mTag BFP]] biobricks into the [[Part:BBa_K2455003|Cell-Penetrating USER Cassette]], the cassette with insertions were expressed in <i>Escherichia coli</i> BL21 under control of the T7 promoter. | ||
| + | |||
| + | [[Image: My_western_image.png|300px|thumb|right|'''Figure 3: Purification of fluorescent proteins.''' Expressed proteins were purified using an Imidazole gradient; E1: 250 mM, E2: 500 mM, E3: 1.0 M, E4: 2.0 M]] | ||
| + | |||
| + | ===Purification=== | ||
| + | Cells were harvested, re-suspended in lysis buffer (25 mM Imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 7.9), and 1.0 mM PMSF), and lysed by sonication. The cell debris was removed by centrifugation at low speed, and the supernatant was transferred to a new falcon tube and mixed with 100 µL [https://www.qiagen.com/us/shop/sample-technologies/protein/expression-purification-detection/ni-nta-agarose/#orderinginformation Ni-NTA Agarose]. The mixture was incubated for one hour at 4 degrees Celcius on a turning table, where after the beads were spun down and the supernatant removed. A series of elution buffers (0.5 M NaCl, 20 mM Tris-HCl; pH 7.9) with increasing Imidazole concentration were used to elute the bound proteins. The elution buffers were added consecutively 100 µl each time followed by centrifuging the sample at low speed, and removing 100 µL eluate. | ||
| + | |||
| + | <b>NOTE:</b> Samples were kept on ice at all times and centrifugation took place at 4 degrees Celsius. | ||
| + | |||
| + | ===SDS-PAGE=== | ||
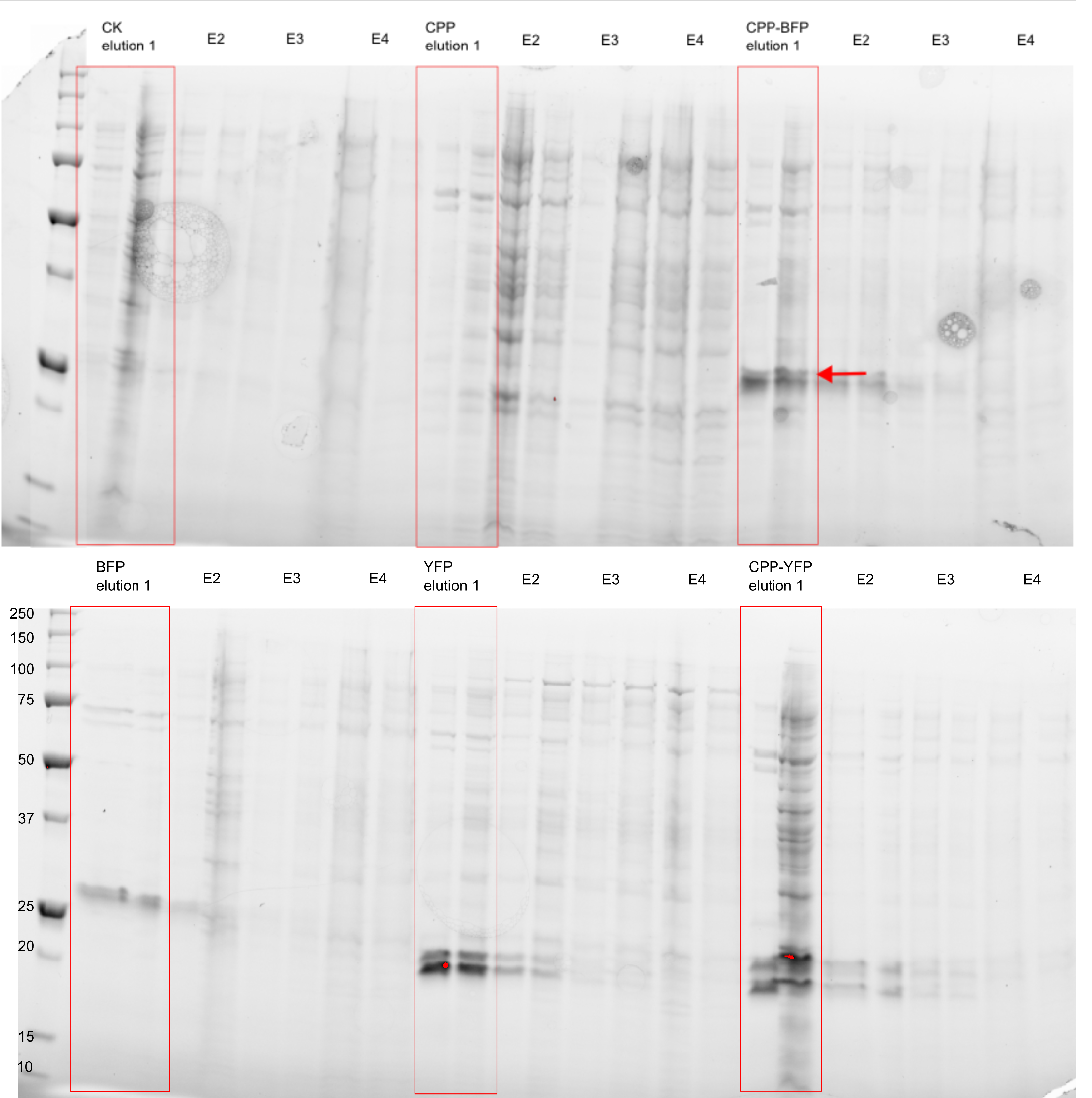
| + | The purity of the samples were analysed by subjecting them to an SDS-PAGE using a [http://www.bio-rad.com/en-cn/product/criterion-precast-gels/criterion-tgx-stain-free-precast-gels pre-cast stain free gel]. In addition to the CPP tagged version of [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K592100|mTag BFP]] two versions without the CPP tag was also purified, along side an empty vector and a vector with an empty [[Part:BBa_K2455003|Cell-Penetrating USER Cassette]]. The Imidazole concentration of the purification ranged from 250 mM to 2.0 M, as can be seen on Figure 3 proteins of the expected size are eluted at Imidazole concentrations of 250 mM for all constructs (BFP & YFP 28kDa, CPP-BFP & CPP-YFP 30kDa). | ||
==Imaging== | ==Imaging== | ||
| − | === | + | All fluorescent imaging was done at 100x magnification using the following setup: |
| + | |||
| + | <b>Yellow fluorescence</b>: | ||
| + | |||
| + | - Excitation time: 250 ms | ||
| + | |||
| + | - Excitation: 460 - 490 nm | ||
| + | |||
| + | - Dichroic Mirror: 505 nm | ||
| + | |||
| + | - Barrier Filter: 510 - 555 nm | ||
| + | |||
| + | |||
| + | <b>Blue fluorescence</b>: | ||
| + | |||
| + | - Excitation time: 100 ms | ||
| + | |||
| + | - Excitation: 330 - 385 nm | ||
| + | |||
| + | - Dichroic Mirror: 400 nm | ||
| + | |||
| + | - Barrier Filter: >420 nm | ||
| + | |||
| + | ===Fluorescent Imaging of Expressing Cells=== | ||
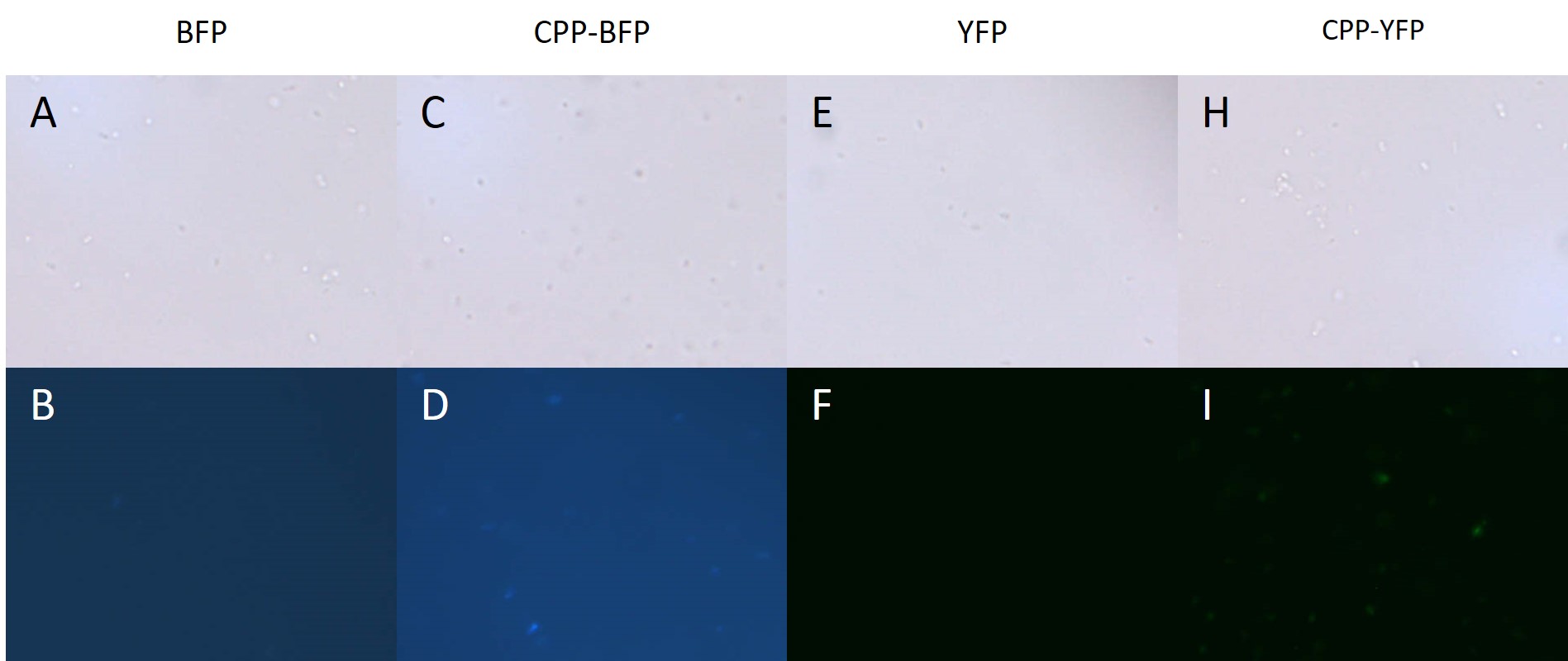
| + | BL21 <i>E. coli</i> expressing either [[Part:BBa_K864100|SYFP2]], [[Part:BBa_K592100|mTag BFP]], [[Part:BBa_K2455002|CPP-SYFP2]], and [[Part:BBa_K2455005|CPP-mTag BFP]] were analysed on a fluorescent microscope at 100x magnification in order to see if the fusion proteins were capable of emitting fluorescent light as their parental counterparts. As can be see on Figure 4 the intensity of the individual cells were as bright with the CPP tag as without. | ||
| + | |||
| + | |||
| + | [[Image: E.coli_export_large.png|600px|thumb|center|'''Figure 4: BL21 <i>E. coli</i> cells expressing either fluorescent proteins.''' A-F) As expected no fluorescence is seen in cells carrying either an empty vector or a vector with an empty [[Part:BBa_K2455003|Cell-Penetrating USER Cassette]]. G+I) Fluorescence of at least equal intensity is seen in cells expressing [[Part:BBa_K2455005|CPP-mTag BFP]] as to those expressing [[Part:BBa_K592100|mTag BFP]]. H+J) Equal fluorescence is seen for cells expressing [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K2455002|CPP-SYFP2]]]] | ||
| + | |||
| + | ===Cell-Penetrating Capabilities of Purified Proteins=== | ||
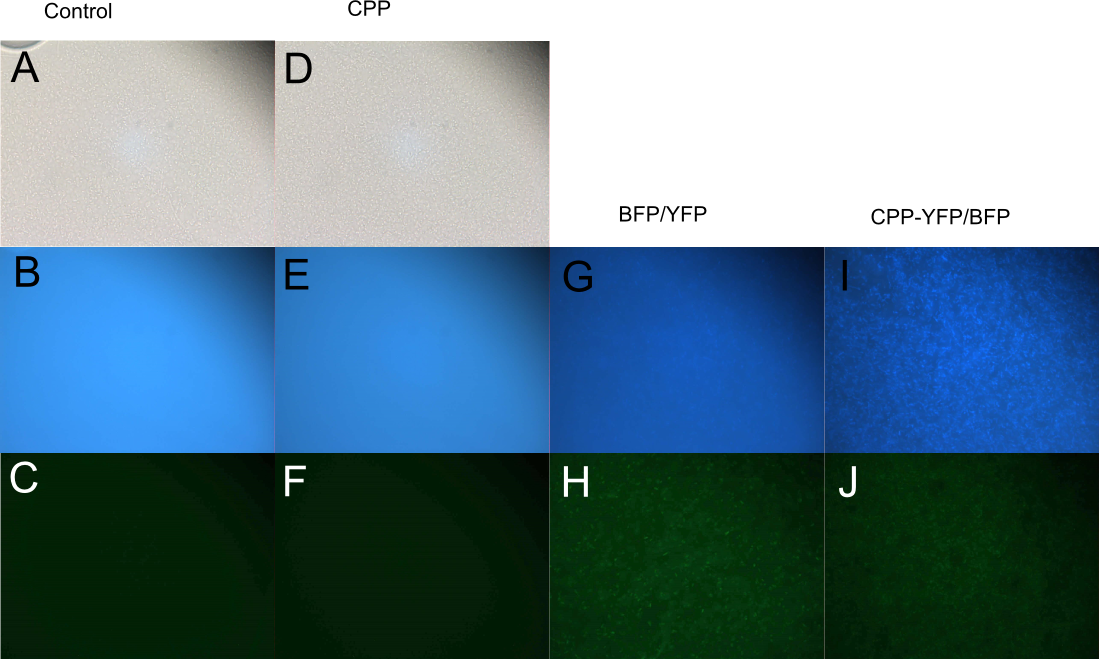
| + | In order to investigate the capabilities of the expressed proteins to enter <i>E. coli</i> cells, 10 µL of <i>E. coli</i> MG1655 overnight culture was mixed with 25 µL purified protein from either Control (empty vector), [[Part:BBa_K864100|SYFP2]], [[Part:BBa_K592100|mTag BFP]], [[Part:BBa_K2455002|CPP]], [[Part:BBa_K2455002|CPP-SYFP2]], or [[Part:BBa_K2455005|CPP-mTag BFP]] and incubated at room temperature for 10 min. Subsequently the cells were pelleted and the supernatant was removed, followed by 2 x wash in 500 mL MQ H<sub>2</sub>O. Finally the cells were re-suspended in 10 µL MQ H<sub>2</sub>O before being imaged on the fluorescent microscope. As can be seen on Figure 5, while neither [[Part:BBa_K864100|SYFP2]] or [[Part:BBa_K592100|mTag BFP]] alone were capable of staining the cells, both [[Part:BBa_K2455002|CPP-SYFP2]] and [[Part:BBa_K2455005|CPP-mTag BFP]] were able too, thus indicating an ability of the [[Part:BBa_K2455003|Cell-Penetrating USER Cassette]] to facilitate internalization of their protein cargo. | ||
| + | |||
| + | |||
| + | [[Image: Slack-imgs.png|700px|thumb|center|'''Figure 5: Fluorescence of <i>E. coli</i> cells treated with fluorescent protein.''' Cells were treated with either [[Part:BBa_K592100|mTag BFP]] (A-B), [[Part:BBa_K2455005|CPP-mTag BFP]] (C-D), [[Part:BBa_K864100|SYFP2]] (E-F), or [[Part:BBa_K2455002|CPP-SYFP2]] (G-H). Cells treated with CPP-tagged protein emitted noticeably more fluorescence.]] | ||
| + | |||
| + | |||
| + | In addition to <i>E. coli</i>, both <i>Pseudomonas putida</i> and <i>Bacillus subtilis</i> were similarly treated with [[Part:BBa_K864100|SYFP2]], [[Part:BBa_K592100|mTag BFP]], [[Part:BBa_K2455002|CPP-SYFP2]], or [[Part:BBa_K2455005|CPP-mTag BFP]] following the same protocol as for <i>E. coli</i> (see above). While <i>B. subtilis</i> showed no signs of protein uptake (data not shown), <i>P. putida</i> showed similar signs to protein uptake as did <i>E. coli</i> (see Fig. 6). | ||
| + | |||
| + | |||
| + | [[Image: Pseudomonas PP.jpg|700px|thumb|center|'''Figure 6: Fluorescence of <i>P. putida</i> cells treated with fluorescent protein.''' Cells were treated with either [[Part:BBa_K592100|mTag BFP]] (A-B), [[Part:BBa_K2455005|CPP-mTag BFP]] (C-D), [[Part:BBa_K864100|SYFP2]] (E-F), or [[Part:BBa_K2455002|CPP-SYFP2]] (G-H). Cells treated with CPP-tagged protein emitted noticeably more fluorescence.]] | ||
Latest revision as of 00:48, 2 November 2017
Cell-Penetrating USER Cassette
This biobrick contains an AsiSI/Nb.BSMI USER Cassette with a N-terminal nona-arginine (R9) Cell-penetrating peptide. The nona-arginine (R9) gives any cassette inserted proteins the ability to pass through plasma membranes. In addition, the USER Cassette carries a N-terminal hexa-histidine tag which allows for easy purification.
For our study we demonstrated that proteins inserted into the cassette can be purified using the histidine-tag, and subsequently used to stain E. coli and Pseudomonas putida cells, presumably through internalization caused by the R9 peptide.
In addition; previous studies have shown that R9 associated proteins, either covalently or non-covalently, were able to enter a variety of cell types (Chang et al., 2005).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Cell-Penetrating Peptides (CPPs) are small peptides, that are able to facilitate transport of a wide variety of cargoes across plasma membranes. CPPs are typically rich in basic residues and originate from viral domains such as the viral HIV tat domain (Eudes & Chugh, 2008). In recent years researchers have tried to make synthetic CPPs, with special interest in peptides constructed solely from arginine residues. The arginine rich sequences have been shown to trigger endocytosis in a wide range of cell types, including onion and potato cells (Chang et al., 2005).
USER Cassette
The biobrick carries the AsiSI/Nb.BsmI USER Cassette which facilitates gene insertion through the use of USER cloning.

For more information on USER cloning see [http://www.cbs.dtu.dk/services/AMUSER/ Amuser] or [http://www.cbs.dtu.dk/services/AMUSER/instructions.php Amuser instructions].
Insertion Requirements
For insertion of genes into the USER Cassette genes have to be amplified with the primers carrying the two following overhangs:
Forward primer: CGTGCGAUCx
Reverse primer: CACGCGAUxx
Where U = Uracil and x = any non-uracil-nucleotide; these are needed to keep the inserted gene in-frame. AsiSI and Nb.BsmI have to be used to facilitate opening of the cassette.
Gene Insertion
In order to test the biobricks ability to facilitate protein import in E. coli we chose to insert two different fluorescent proteins into the biobrick, SYFP2 and mTag BFP.
Opening of USER Cassette
To facilitate insertion, we first had to prepare the vector, this was done in a two-step digestion reaction. First to open the cassette 42 µL purified vector was mixed with 2.5 µL AsiSI, 10 µL 10x Tango Buffer, and 45.5 µL Nuclease-free water. The mixture was then incubated for 3 hours at 37 degrees Celsius. Then we ran 5 µL of the linearised vector for 20 min at 100 V on a 1 % Agarose gel to check for proper linearization. As shown on Figure 2 the vector goes from a smaller band, typical for circular (supercoiled) pieces of DNA, while the digested vector has a band consistent with the full 6kbp of the vector.
Following digestion the vector was subjected to column-purified using [http://omegabiotek.com/store/product/e-z-n-a-gel-extraction-kit/ E.Z.N.A.® Gel Extraction Kit] and [http://omegabiotek.com/store/wp-content/uploads/2013/05/D2500.D2501-Gel-Extraction-Kit-Combo-Online.pdf standard protocol] for gel-extraction with 150 µL binding buffer being added in the first step.
Finally the vector was nicked by mixing the complete 50 µL elute from the column-purification with 1 µL Nb.BsmI, 6 µL 10x NEBuffer 3.1, and 3 µL Nuclease-free water. The mixture was incubated for 3 hours at 65 degrees Celcius.
Gene Amplification
In order to facilitate the insertion of the SYFP2 and mTag BFP biobricks into the USER Cassette, the two genes were amplified using the following sets of primers in order to add the appropriate overhangs:
SYFP2:
Forward primer: CGTGCGAUCA-ATGGTTAGCAAGGGCGAAG
Reverse primer: CACGCGAUGA-TTTATACAGCTCATCCATACCCAGGG
mTag BFP:
Forward primer: CGTGCGAUCA-ATGAGCGAACTGATCAAAGAG
Reverse primer: CACGCGAUGA-ATTCAGTTTATGACCCAGC
And gel-extracted using [http://omegabiotek.com/store/product/e-z-n-a-gel-extraction-kit/ E.Z.N.A.® Gel Extraction Kit] and [http://omegabiotek.com/store/wp-content/uploads/2013/05/D2500.D2501-Gel-Extraction-Kit-Combo-Online.pdf standard protocol].
Insertion of BFP and YFP
To insert the gel-extracted PCR products 1 µL of the PCR product was mixed with 2 µL linearised vector, 0.5 µL CutSmart® Buffer, 0.5 µL USER® Enzyme, and 1 µL MQ H2O. The mixture was incubated in a PCR machine for 25 min at 37 degrees Celcius, 20 min at 25 degrees Celcius, 10 min at 20 degrees Celcius, and 10 min at 15 degrees Celcius.
The insertions were validated through PCR amplification of the inserts followed by sequencing.
Purification and SDS-PAGE
Following the validated insertion of the SYFP2 and mTag BFP biobricks into the Cell-Penetrating USER Cassette, the cassette with insertions were expressed in Escherichia coli BL21 under control of the T7 promoter.
Purification
Cells were harvested, re-suspended in lysis buffer (25 mM Imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 7.9), and 1.0 mM PMSF), and lysed by sonication. The cell debris was removed by centrifugation at low speed, and the supernatant was transferred to a new falcon tube and mixed with 100 µL Ni-NTA Agarose. The mixture was incubated for one hour at 4 degrees Celcius on a turning table, where after the beads were spun down and the supernatant removed. A series of elution buffers (0.5 M NaCl, 20 mM Tris-HCl; pH 7.9) with increasing Imidazole concentration were used to elute the bound proteins. The elution buffers were added consecutively 100 µl each time followed by centrifuging the sample at low speed, and removing 100 µL eluate.
NOTE: Samples were kept on ice at all times and centrifugation took place at 4 degrees Celsius.
SDS-PAGE
The purity of the samples were analysed by subjecting them to an SDS-PAGE using a [http://www.bio-rad.com/en-cn/product/criterion-precast-gels/criterion-tgx-stain-free-precast-gels pre-cast stain free gel]. In addition to the CPP tagged version of SYFP2 and mTag BFP two versions without the CPP tag was also purified, along side an empty vector and a vector with an empty Cell-Penetrating USER Cassette. The Imidazole concentration of the purification ranged from 250 mM to 2.0 M, as can be seen on Figure 3 proteins of the expected size are eluted at Imidazole concentrations of 250 mM for all constructs (BFP & YFP 28kDa, CPP-BFP & CPP-YFP 30kDa).
Imaging
All fluorescent imaging was done at 100x magnification using the following setup:
Yellow fluorescence:
- Excitation time: 250 ms
- Excitation: 460 - 490 nm
- Dichroic Mirror: 505 nm
- Barrier Filter: 510 - 555 nm
Blue fluorescence:
- Excitation time: 100 ms
- Excitation: 330 - 385 nm
- Dichroic Mirror: 400 nm
- Barrier Filter: >420 nm
Fluorescent Imaging of Expressing Cells
BL21 E. coli expressing either SYFP2, mTag BFP, CPP-SYFP2, and CPP-mTag BFP were analysed on a fluorescent microscope at 100x magnification in order to see if the fusion proteins were capable of emitting fluorescent light as their parental counterparts. As can be see on Figure 4 the intensity of the individual cells were as bright with the CPP tag as without.
Cell-Penetrating Capabilities of Purified Proteins
In order to investigate the capabilities of the expressed proteins to enter E. coli cells, 10 µL of E. coli MG1655 overnight culture was mixed with 25 µL purified protein from either Control (empty vector), SYFP2, mTag BFP, CPP, CPP-SYFP2, or CPP-mTag BFP and incubated at room temperature for 10 min. Subsequently the cells were pelleted and the supernatant was removed, followed by 2 x wash in 500 mL MQ H2O. Finally the cells were re-suspended in 10 µL MQ H2O before being imaged on the fluorescent microscope. As can be seen on Figure 5, while neither SYFP2 or mTag BFP alone were capable of staining the cells, both CPP-SYFP2 and CPP-mTag BFP were able too, thus indicating an ability of the Cell-Penetrating USER Cassette to facilitate internalization of their protein cargo.
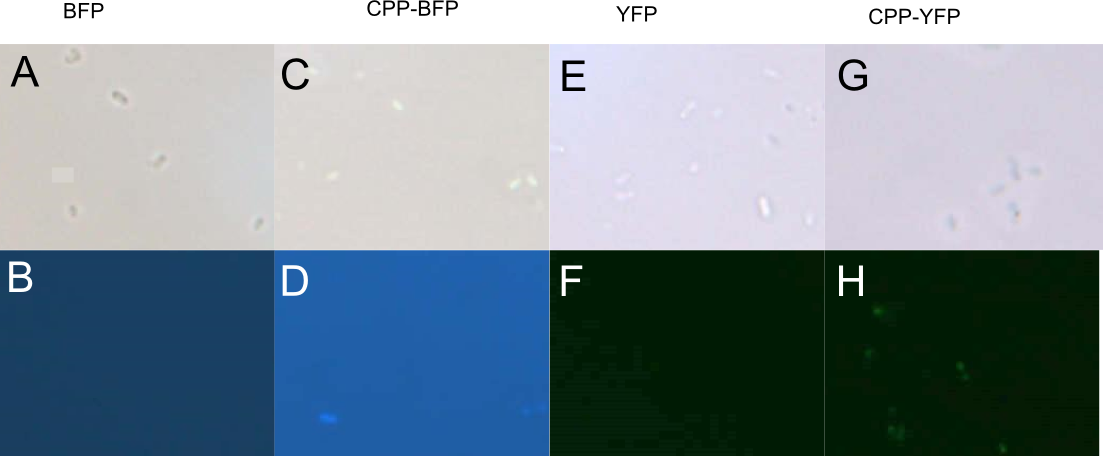
In addition to E. coli, both Pseudomonas putida and Bacillus subtilis were similarly treated with SYFP2, mTag BFP, CPP-SYFP2, or CPP-mTag BFP following the same protocol as for E. coli (see above). While B. subtilis showed no signs of protein uptake (data not shown), P. putida showed similar signs to protein uptake as did E. coli (see Fig. 6).