Difference between revisions of "Part:BBa K1758313"
(→Usage and Biology) |
(→Usage and Biology) |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <partinfo>BBa_K1758313 short</partinfo> | ||
| + | |||
| + | Repressor for chromium induceble promoter crProm under the control of constitutive promoter (K608002) | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
<html> | <html> | ||
<p align="justifiy"> | <p align="justifiy"> | ||
| − | Our sensor for chromium detection consists of <i>chrB</i> the repressor and the chromate specific promoter <i>chrP</i>. The promoter | + | Our sensor for chromium detection consists of <i>chrB</i> the repressor and the chromate specific promoter <i>chrP</i>. The promoter <i>chrP</i> is regulated by the repressor combined they are used for <i>in vivo</i> characterization </figcaption> |
</figure>, which binds Cr-ions. Behind the promoter is a <i>sfGFP</i> for detection of a fluorescence signal.</p> | </figure>, which binds Cr-ions. Behind the promoter is a <i>sfGFP</i> for detection of a fluorescence signal.</p> | ||
<i>In vivo</i> we could show that the addition of different concentrations of chromium have different effects to transcription of <i>sfGFP</i>.</p> | <i>In vivo</i> we could show that the addition of different concentrations of chromium have different effects to transcription of <i>sfGFP</i>.</p> | ||
| Line 19: | Line 23: | ||
<html> | <html> | ||
<body style="width:600px;"> | <body style="width:600px;"> | ||
| + | |||
| + | <h2><i>in vivo</i> characterisation</h2> | ||
<p align="justify"> | <p align="justify"> | ||
Our sensors were cultivated in the BioLector. Due to the accuracy of this device we could measure our sample in duplicates.</br></br> | Our sensors were cultivated in the BioLector. Due to the accuracy of this device we could measure our sample in duplicates.</br></br> | ||
| Line 32: | Line 38: | ||
</br><p>Our data lead to the conclusion that in a cell based system it is possible to detect chromium. | </br><p>Our data lead to the conclusion that in a cell based system it is possible to detect chromium. | ||
In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation. </p> | In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation. </p> | ||
| + | |||
| + | <h2><i>in vitro</i> characterisation</h2> | ||
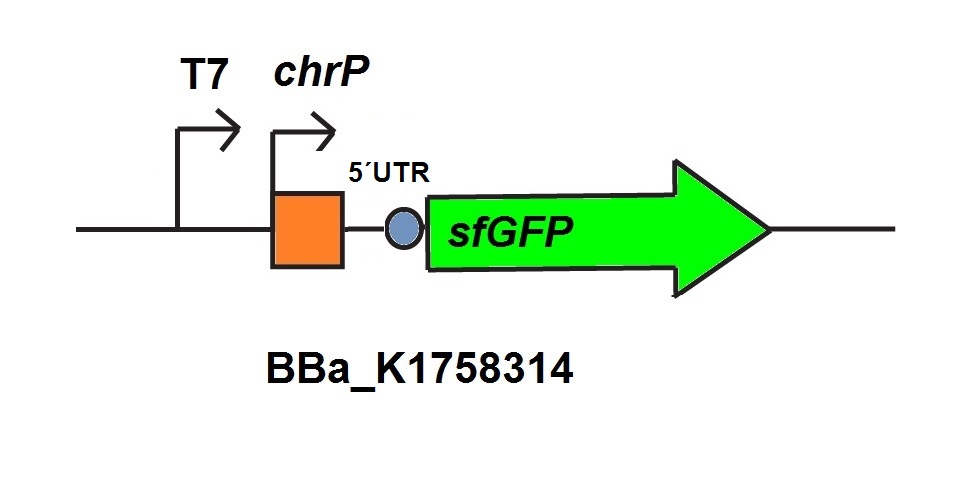
| + | <p>For the characterization of the chromium sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract produced from cells which contain the plasmid (<a href="https://parts.igem.org/Part:BBa_K1758310" target="_blank">BBa_K1758310</a>). The plasmid contains the gene <i>chrB</i> under the control of a constitutive promoter, so that the cell extract is enriched with repressor molecules. In addition to that we added plasmid-DNA of the chromium specific promoter <i>chrP</i> with 5’UTR-sfGFP under the control of T7-promoter (<a href="https://parts.igem.org/Part:BBa_K1758314" target="_blank">BBa_K1758314</a>)to the cell extract. The T7-promoter is needed to get a better fluorescence expression. </p> | ||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <figure> | ||
| + | <a><img src=" https://static.igem.org/mediawiki/2015/e/e4/Bielefeld-CeBiTec_in_vitro_ChrB-part.jpeg" alt="repressor construct used for in vivo characterisation" width="500px"></a> <figcaption> To produce the cell extract for <i>in vitro</i> characterization a construct (<a href="https://parts.igem.org/Part:BBa_K1758310" target="_blank">BBa_K1758310</a> ) with chromium repressor under the control of a constitutive promoter and strong RBS (BBa_K608002) is needed.</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <figure> | ||
| + | <a><img src=" https://static.igem.org/mediawiki/2015/1/1f/Bielefeld-CebiTec_in_vitro_T7-chrP-UTR-sfGFP.jpeg" alt="promoter construct used for in vivo characterisation" width= "500px"></a> <figcaption> T7-chrP-UTR-sfGFP <a href="https://parts.igem.org/Part:BBa_K1758314" target="_blank">BBa_K1758314</a> used for <i>in vitro</i> characterization.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <figure > | ||
| + | <a><img src="https://static.igem.org/mediawiki/2015/9/99/Bielefeld-CeBiTec_Influence_of_chromium_on_the_cell_extract.jpeg" alt="Adjusting the detection limit" width= "800px"></a> | ||
| + | <figcaption>Influence of different chromium concentrations on our crude cell extract. Error bars represent the standard deviation of three biological replicates. </figcaption> | ||
| + | </figure> | ||
| + | |||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 12:04, 20 September 2015
Chromium repressor under control of constitutive promoter and strong RBS,chromium responsive promote
Repressor for chromium induceble promoter crProm under the control of constitutive promoter (K608002)
Usage and Biology
Our sensor for chromium detection consists of chrB the repressor and the chromate specific promoter chrP. The promoter chrP is regulated by the repressor combined they are used for in vivo characterization , which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal.
In vivo we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP. e used this repressor for our chromium sensor. Originaly its from Ochrobactrum triti ci5bvl1. We codon optimized it for use in E.coli. It is essential for our chromium sensor device BBa_K1758310. In combination with BBa_K1758312,BBa_K1758314 This part is used as our in vivo cerates our chromium sensor.Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 55
Illegal NheI site found at 966
Illegal NheI site found at 989 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 124
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1237
Results
in vivo characterisation
Our sensors were cultivated in the BioLector. Due to the accuracy of this device we could measure our sample in duplicates.


Our data lead to the conclusion that in a cell based system it is possible to detect chromium. In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation.
in vitro characterisation
For the characterization of the chromium sensor with CFPS we used parts differing from that we used in vivo characterization. For the in vitro characterization we used a cell extract produced from cells which contain the plasmid (BBa_K1758310). The plasmid contains the gene chrB under the control of a constitutive promoter, so that the cell extract is enriched with repressor molecules. In addition to that we added plasmid-DNA of the chromium specific promoter chrP with 5’UTR-sfGFP under the control of T7-promoter (BBa_K1758314)to the cell extract. The T7-promoter is needed to get a better fluorescence expression.