Part:BBa_K2446042
ZF_54-8_KRAB
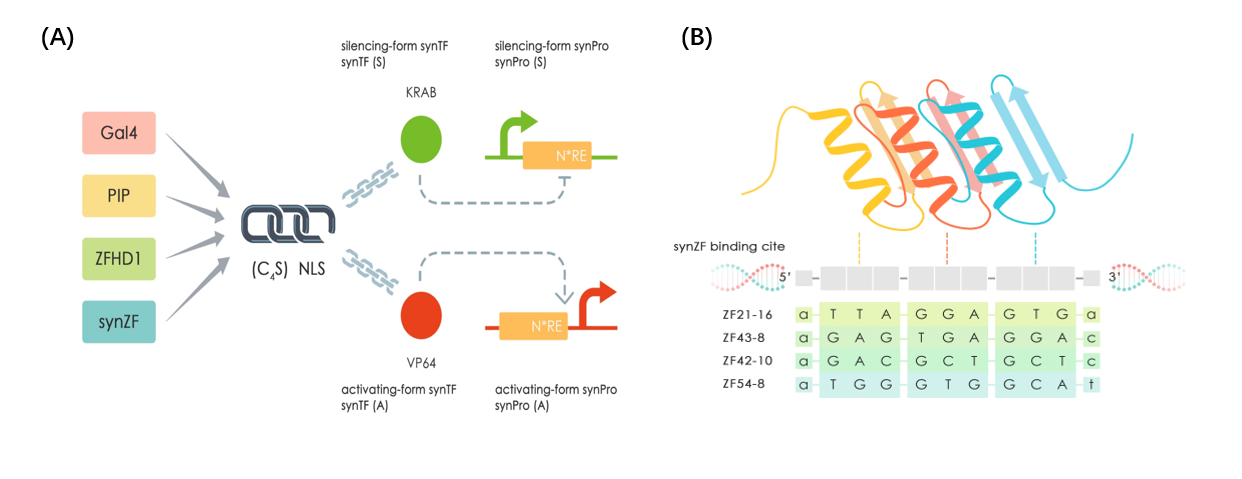
This part is one of our mammalian synthetic transcription factors (SynTFs) based on Cys2-His2 zinc finger 54-8 (ZF_54-8) as protein chassis. [1]. ZF_54-8_KRAB (or TF-KRAB-6 in our project) containing three core domains from N-terminal to C-terminal: ZF_54-8 DNA binding domain, nuclear location sequence and KRAB transcription regulating domain [2].And a (G4S) linker was added between DBD and NLS for providing region flexibility [3]. ZF_54-8-DBD enable binding to specific DNA sequences(as figure 1 shows), so that we can use ZF_54-8-KRAB as a specific transcription factors to repress the expression of our mammalian synthetic promoter SV40-8_ZF_54-8 (BBa_K2446029).
The information of other SynTF-SynPros is showed in the table below.
| SynTFs | SynPros |
|---|---|
| Gal4-KRAB(TF-KRAB-1) BBa_K2446037 | Sv40-UAS(Sv40-UAS) BBa_K2446036 |
| ZF_PIP_KRAB(TF-KRAB-2) BBa_K2446045 | SV40_2/4/8_PIP BBa_K2446033/BBa_K2446034/BBa_K2446035 |
| ZF_21-16KRAB(TF-KRAB-3) BBa_K2446039 | SV40_8_ZF_21-16 BBa_K2446030 |
| ZF_42-10_KRAB(TF-KRAB-4) BBa_K2446040 | SV40_8_ZF_42-10 BBa_K2446025 |
| ZF_43-8_KRAB(TF-KRAB-5) BBa_K2446041 | SV40_2/4/8_ZF_43-8 BBa_K2446026/BBa_K2446027/BBa_K2446028 |
| ZF_54-8_KRAB(TF-KRAB-6) BBa_K2446042 | SV40_8_ZF_54-8 BBa_K2446029 |
| ZFHD1_KRAB(TF-KRAB-7) BBa_K2446043 | SV40_4_ZFHD1 BBa_K2446032 |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 101
- 1000COMPATIBLE WITH RFC[1000]
Experiments
SynTF-SynPro Pairs
Figure 2: the testing circuits of ZF_54-8-KRAB& Sv40-8_ ZF_54-8 pair
To make sure the SynTF-SynPro pairs work in mammalian cells, we use the circuits above to test if the ZF_54-8-KRAB can repress the expression of Sv40-8_ZF_54-8 indeed. ZF_54-8-KRAB is linked to the C terminal of EGFP by the link of P2A. And mCherry expressions is controlled by corresponding SynPro (Sv40-8_ ZF_54-8). These circuits are both inserted in to the mammalian expression vactor pML2. We transfect pML2-Sv40-8_ ZF_54-8 into Hela cells and measure the fluorescence intensity of mCherry by flow cytometer to get the basic expression intensity of Sv40_ ZF_54-8. We also co-transfect the pML2- ZF_54-8-KRAB with pML2-Sv40-8_ ZF_54-8 into Hela cells at the same time. Then measure the fluorescence intensity of mCherry again to get the expression intensity of Sv40-8_ZF_54-8 influenced by ZF_54-8-KRAB. The results of the experiment is showed below. The expression intensity of Sv40-54-8 is too low to detect the silencing rate of ZF_54-8-KRAB.
Figure 3:The results of ZF_54-8-KRAB&SV40_ ZF_54-8 testing: (A) The red points is the cells before co-transfecting ZF_54-8-KRAB and the blue points is the cells after co-transfecting ZF_54-8-KRAB. It’s easy to see that the red points depart from the diagonal and higher than the blue pints. So the expression of mCherry silenced after the expression of ZF_54-8-KRAB;(B) The red area is the fluorescence intensity of mCherry before co-transfecting ZF_54-8-KRAB and the blue area is the intensity after co-transfecting ZF_54-8-KRAB.(C) The statistical result of all of the SynTFs-SynPros pairs: The expression intensity of Sv40-54-8 is too low to detect the silencing rate of ZF_54-8-KRAB.
SynTF-SynPro Orthogonality
To construct our [http://2017.igem.org/Team:Fudan/Model/GTN| Strip module], more than one SynTF-SynPro pairs would be applied. Thus, the interaction between the pairs would influence or ruin our construction. We did massive orthogonality experiments to avoid that. We observed all of the 5 pairs were actually orthogonal, as you could see the grids on the diagonal were always the darkest. The three DBDs commonly used in previous works were didn’t let us down. However, the expression level of these RE loaded SynPros were relative low compared to SynPro(S)-ZF serials. As the blue rectangle in the lower right corner of the orthogonality may showed the SynPro(S)-ZF has high basic expression with unpaired SynTFs, but could be silenced to the similar fold of commonly used DBDs corresponding SynPros. The SynPro(S)-ZF was likely won’t be target by other unpaired DBD, hence the purple appeared on the bottom rows.
Figure 4: the SynTF-SynPro pairs’ Orthogonality. (A) SynPro(S)-54-8;8* were constantly silenced.; (B)Grids in blue rectangle showed that SynTF-SynPro pairs constructed by using SynZF as DBD with well orthogonality. Grids in pink rectangles replaced our favorite SynTF-SynPro pairs. At least 20,000 cells were analyzed for each condition in both histogram and each grid in heat map. Data are recorded by FACS at 24h after cotransfecting.
Reference
[1] Lohmueller, J. J., Armel, T. Z. & Silver, P. A. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Research 40, 5180--5187 (2012).
[2] Witzgall, R., O'Leary, E., Leaf, A., Onaldi, D. & Bonventre, J. V. The Krüppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proceedings of the National Academy of Sciences 91, 4514-4518 (1994).
[3] Chen, X., Zaro, J. L. & Shen, W.-C. Fusion protein linkers: property, design and functionality. Advanced drug delivery reviews 65, 1357-1369 (2013).| None |