Part:BBa_K1073035
Inducible ampR expression combined with RhlR, LasI and eforRed expression cassettes
This device is intended to be used in concert with BBa_K1073034. Using both BBa_K1073034 and BBa_K1073035 a synthetic consortia between two strains can be created when they are incubated in ampicillin containing medium. The two strains depend on each other and can only grow when the other is present in the culture broth.
BBa_K1073035 is a combination of BBa_K1073028 and BBa_K1073032 and includes parts of the rhl and las quorum sensing system of Pseudomonas aeruginosa. It includes constitutive expression cassettes for RhlR, LasI and eforRed.
The expression of ampR is regulated by the promoter of the rhl system. In order to induce ampR expression the transcription regulator RhlR and the specific autoinducer molecule N-butyryl homoserine lactone have to be present. N-butyryl-HSL is synthesized by RhlI (this protein is not included in the construct). However, N-butyryl-HSL can be added to the culture broth. RhlR and N-butyryl-HSL build a complex that binds to specific sequences of the promoter region and induces expression of the downstream gene (in this case ampR) [Medina et al., 2003].
LasI synthesizes the specific autoinducer of the las quorum sensing system N-3-oxododecanoyl homoserine lactone. The autoinducer molecules are secreted into the medium.
The expression of eforRed can be used as selection marker. EforRed exhibits a strong pink color when expressed and is visible to the naked eye in less than 24 h during incubation on agar plates or in liquid culture.
Usage and Biology
Two different strains containing BBa_K1073034 and BBa_K1073035 respectively will together synthesize all molecules required to induce the promoters of the rhl and las system and thus activating the expression of ampRof one another.
Possible Applications
Using both BBa_K1073035 and BBa_K1073034 a synthetic consortia between two strains can be created when they are incubated in ampicillin containing medium. The two strains depend on each other and can only grow when the other is present in the culture broth.
It can also be used without BBa_K1073034 when synthetic N-butyryl-HSL is added to the culture broth.
Growth Characteristics
This device was tested in E. coli Top10 F' . During batch cultivation in complex medium containing ampicillin growth of cells is inhibited unless autoinducer molecules N-butyryl-HSL (in this case synthetic N-butyryl-HSL) are added. Background activity of the beta lactamase due to the leakiness of the rhl promoter causes a slow degredation of ampicillin so that cells start to grow after a Long extented lag-phase.
iGEM Team Braunschweig 2013: Growth curve of E. coli Top10 F' containing BBa_K1073035 on the high copy plamsid pSB1C3. Comparison of growth in ampicillin containing complex medium in presence and absence of rhl autoinducer N-butyryl-HSL.
Induction of ampR expression by N-butyryl-HSL
Growth of cells containing BBa_K1073035 can be induced by synthetic autoinducer (see Growth Characteristics) and also by the supernatant of a culture of cells containing BBa_K1073034. A piece of filter paper was soaked with the culture's supernatant and placed on top of a 2xYT agar plate containing ampicillin. Growth of cells containing BBa_K1073035 could only be observed close to the filter. This not only proves the inducibility of this contruct but also the production of N-butyryl-HSL by RhlI encoded in BBa_K1073034.
iGEM Team Braunschweig 2013: Growth of E. coli Top10 F' containing BBa_K1073035 on the high copy plasmid pSB1C3 induced by the supernatant of expressed BBa_K1073034. The supernatant contains N-butyryl-HSL produced by RhlI.
Production of las autoinducer N-3-oxododecanoyl-HSL
In order to verify the production of las autoinducer N-3-oxododecanoyl-HSL by the autoinducer synthase LasI encoded in this device a E. coli JM109 bioluminescence reporter strain was used which contains a lasRI'::luxCDABE cassette on a pSB1075 plasmid backbone [Winson et al., 1998]. The presence of N-3-oxododecanoyl-HSL in the medium leads to the expression of luxCDABE . E. coli JM109::lasRI'::luxCDABE was incubated with diluted supernatant of a culture of cells containing BBa_1073035. Bioluminescence was measured by using a plate reader.
iGEM Team Braunschweig 2013: Measurement of bioluminescence produced by reporter strain E. coli JM109::lasRI'::luxCDABE due to the presence of N-3-oxododecanoyl-HSL produced by LasI in the medium.
Expression of chromoprotein eforRed
This device encodes the eforRed expression cassette (see BBa_K1073022). When expressed this chromoprotein exhibits a pink color and therefore can be used as a selection marker for cells containing BBa_K1073035. The color is visible to naked eye in less than 24 h on agar plate and in liquid culture.
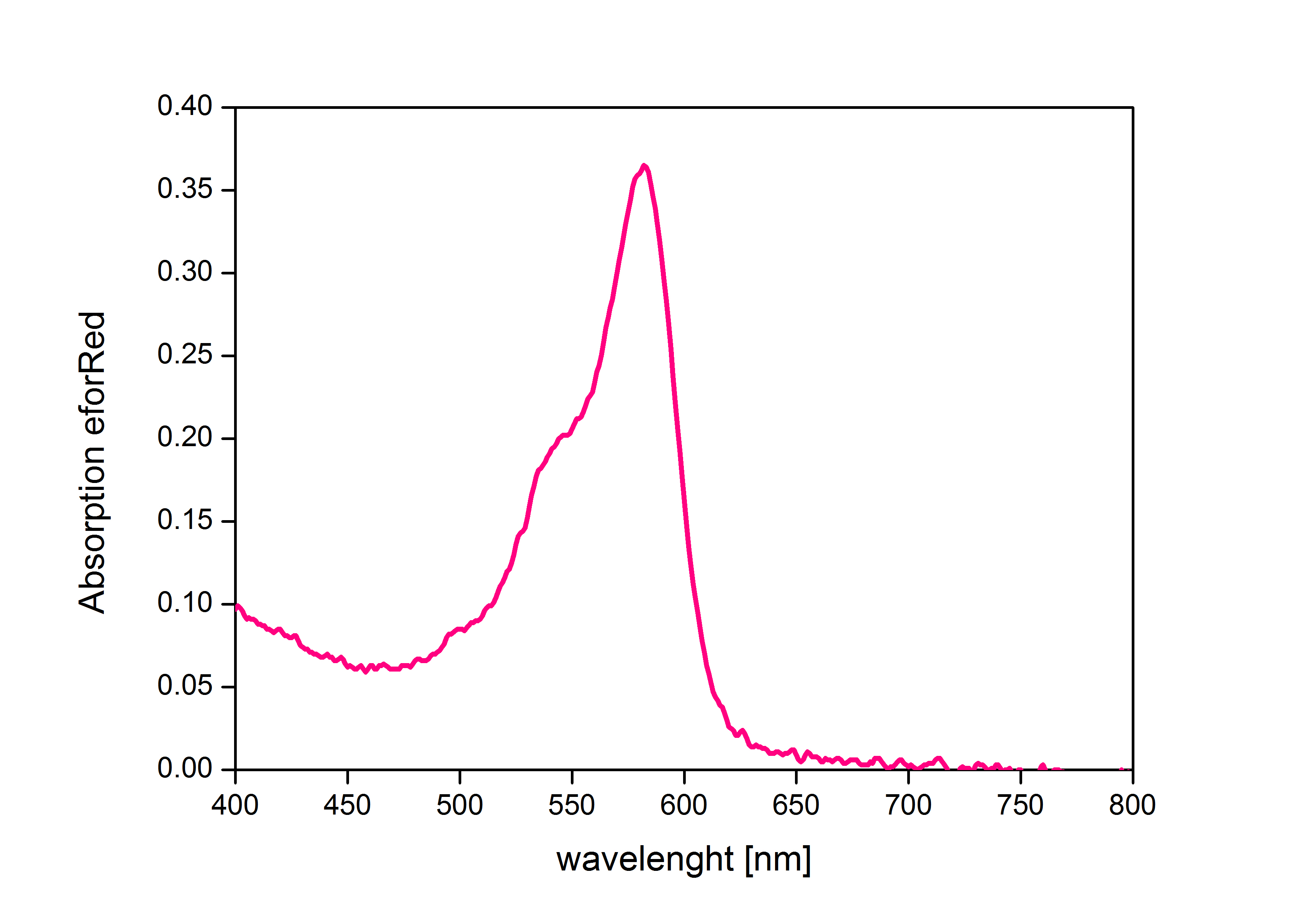
In order to avoid absorption by eforRed during OD measurement of a bacterial culture, the absorption spectrum was measured. An optimal wavelength for spectral optical density measurement was determined to be at 520nm according to this spectrum.
iGEM Team Braunschweig 2013:
Left: Cell pellet and liquid culture (2xYT medium) of cells containing BBa_K1073035. Cells turn red due to the constitutive expression of the chromoprotein eforRed encoded in this device.
Right: Absorption spectrum of chromoprotein eforRed in a wavelength range of 400-800 nm.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1096
Illegal NheI site found at 1119
Illegal NheI site found at 2784
Illegal NheI site found at 2807 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2626
Illegal BamHI site found at 1391 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2188
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1866
| None |