Recombination/Bacteriophage P1-derived Cre-lox
< Back to Recombination systems, DNA recombination sites, or Recombinases
| Chris Anderson, a professor of bioengineering at UC Berkeley, constructed the lox recombination site BBa_J61046. | Eimad Shotar, a member of the [http://2007.igem.org/Paris 2007 Paris iGEM team], constructed the lox recombination sites BBa_I718016 and BBa_I718017. |
The following text is excerpted from Siegel et al. Siegel04.
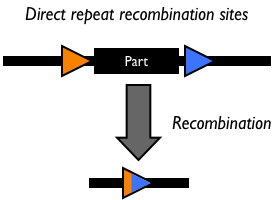
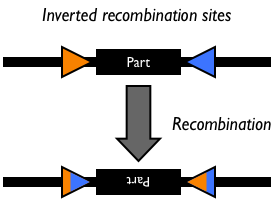
Bacteriophage P1 uses a site-specific recombination system that is responsible for partitioning newly synthesized genomic copies during replication Abremski, Hoess. This system is composed of a 38-kD phage-encoded Cre recombinase that mediates symmetrical recombination between two 34-bp loxP sites Abremski, which are recreated after recombination. Recombination between two compatible loxP sites will excise or invert the intervening DNA in the case of an intramolecular reaction or transfer suitably flanked loxP DNA in an intermolecular double cross-over recombination event. The Cre/loxP system does not require accessory factors to carry out recombination in vivo or in vitro. Importantly, the Cre/lox system has also been shown to be functional in site-specific recombination in mammalian cell lines Sauer88.
Studies have also identified several hetero-specific loxP sequences that exclusively recombine with themselves, but not with wild-type lox Hoess86, Sauer92, Lee98, Siegel01. For example, the mutant lox66 and lox71 sites make the inversion reaction largely unidirectional, so that the intervening DNA does not flip back to the original orientation.
Recombination sites
| Name | Description | Sequence | Recombinase | Length |
|---|---|---|---|---|
| BBa_I718016 | lox66 | . . . cttggtatagcatacattatacgaacggta | 34 | |
| BBa_I718017 | lox71 | . . . gttcgtatacgatacattatacgaagttat | 34 | |
| BBa_J61046 | [Lox] site for recombination | . . . cttcgtataatgtatgctatacgaagttat | 34 | |
| BBa_K1680005 | loxP Site | . . . cttcgtatagcatacattatacgaagttat | 34 | |
| BBa_K315011 | Variant reverse lox N | . . . cttcgtatagtataccttatacgaagttat | 34 | |
| BBa_K416002 | 36 Base Pair LoxP | . . . tcgtataatgtatgctatacgaagttatcg | 36 | |
| BBa_K886000 | Fixed lox71 | . . . gttcgtatagcatacattatacgaagttat | 34 |
Recombinases
| Name | Protein | Description | Direction | KEGG | UniProt | E.C. | Recombination site | Length |
|---|---|---|---|---|---|---|---|---|
| BBa_K1680008 | Δ1-19 Cre recombinase | 972 | ||||||
| BBa_K3120003 | A human codon-optimized SpCas9 | 4101 | ||||||
| BBa_K112115 | a~Cre | 1036 | ||||||
| BBa_I716212 | Cre (GTG start) | 1032 | ||||||
| BBa_I716213 | Cre (TTG start) | 1032 | ||||||
| BBa_J61047 | Cre DNA recombinase | 1037 | ||||||
| BBa_K1680007 | Cre recombinase | 1029 | ||||||
| BBa_K112122 | Cre with stop codon | 1032 | ||||||
| BBa_K1680017 | Cre-Dronpa Fusion | 1728 | ||||||
| BBa_K1680018 | Cre-Dronpa fusion | 1746 | ||||||
| BBa_K1680019 | Cre-dronpa fusion | 1764 | ||||||
| BBa_K1680020 | Cre-dronpa fusion | 1782 | ||||||
| BBa_K1680021 | Dronpa caged Cre with NLS | 2433 | ||||||
| BBa_K1680022 | Dronpa caged Cre with NLS | 2451 | ||||||
| BBa_K1680023 | Dronpa caged Cre with NLS | 2469 | ||||||
| BBa_K1680024 | Dronpa caged Cre with NLS | 2487 |
Composite parts
| Name | Type | Description | Length | Status |
|---|---|---|---|---|
| BBa_I718008 | Generator | araC-pBad-rbs-cre | 2276 | In stock |
| BBa_K082011 | Translational_Unit | B0034-J61047 | 1055 | It's complicated |
| BBa_K132027 | Generator | cre induced by iptg | 2781 | It's complicated |
| BBa_K132028 | Generator | cre terminator | 1195 | It's complicated |
| BBa_K5331011 | Plasmid | fl4a-nat-loxp | 673 | |
| BBa_I718007 | Translational_Unit | RBS-CRE | 1058 | In stock |
| BBa_K886003 | Device | Recombination Device composed by Cre-lox71 | 1079 | It's complicated |
| BBa_K886001 | Device | Recombination Device composed by lox71-Cre recombinase | 1077 | In stock |
| BBa_K1680025 | Reporter | RFP-luciferase Cre reporter | 2900 | It's complicated |
References
<biblio>
- Abremski pmid=6319400
- Hoess pmid=6230671
- Hamilton pmid=6333513
- Hoess86 pmid=3457367
- Sauer88 pmid=2839833
- Sauer92 pmid=1554399
- Lee98 pmid=9714735
- Sauer98 pmid=9608509
- Siegel01 pmid=11576551
- Sauer02 pmid=12624421
- Siegel04 pmid=15173117
</biblio>