Part:J61101
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656012 which is this part adapted to the Golden Gate technology.
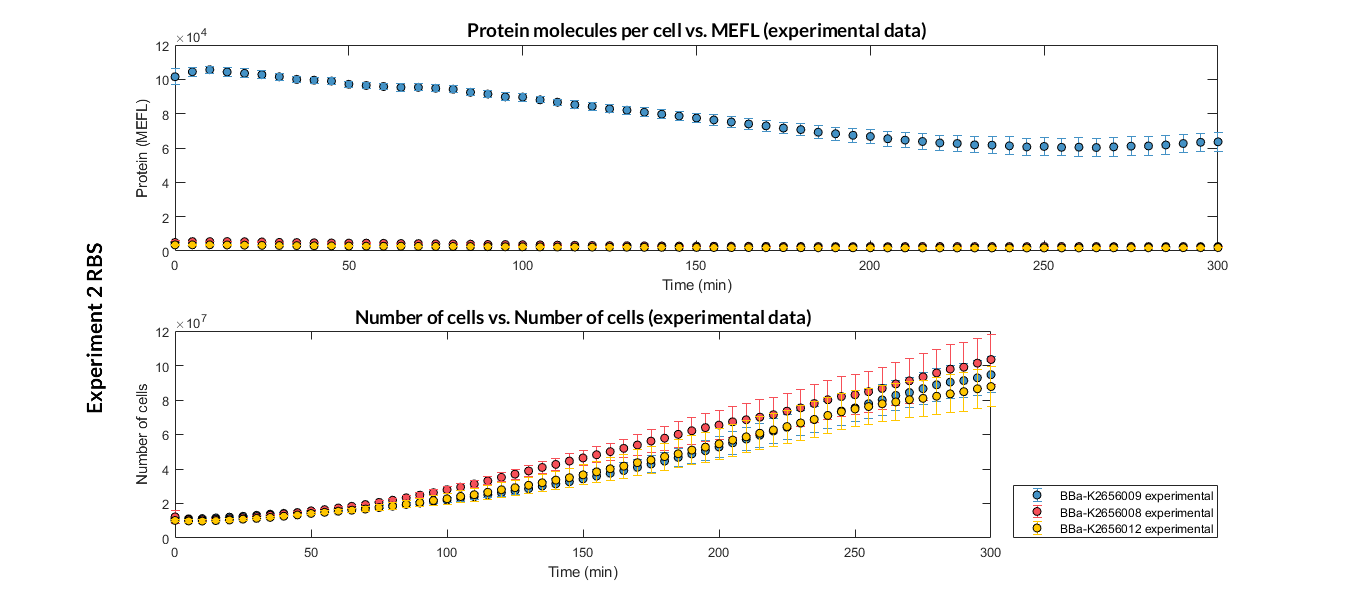
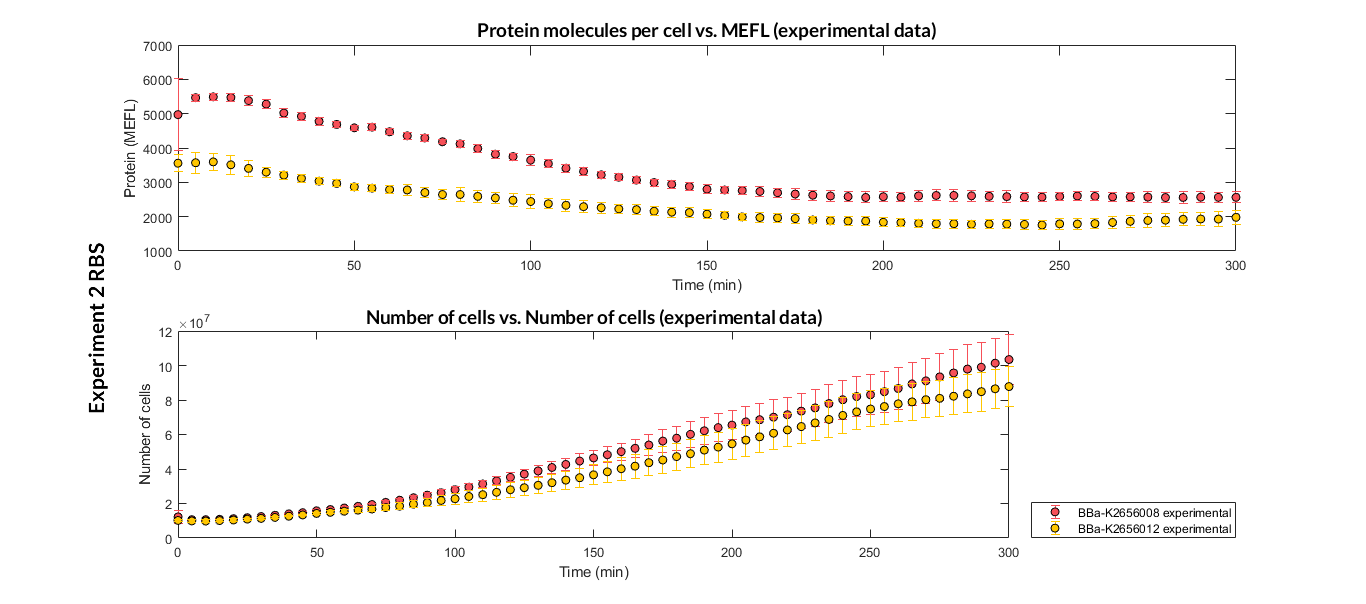
Characterization of the this part was performed with the transcriptional unit BBa_K2656104, which was used in a comparative RBS expression experiment with composite parts BBa_K265610 and BBa_K2656101. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS.
| Table 1. Optimized parameters for the TU with BBa_K2656008 RBS. | |||
| Parameter | Value | ||
| Translation rate p | p = 0.02889 min-1 | ||
| Dilution rate μ | μ = 0.0118 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.032 | ||
| p parameter ratio (pRBS/pref) | 0.032 | ||
//ribosome/prokaryote/ecoli
//chassis/prokaryote/ecoli
//direction/forward
//regulation/constitutive
| None |

 1 Registry Star
1 Registry Star