Part:BBa_K808000
araC-Pbad - Arabinose inducible regulatory promoter/repressor unit
This part contains the promoter as well as the coding sequence for the repressor AraC which is transcribed in the opposite direction. (“upstream”) By binding to L(+)-arabinose, AraC changes its conformation. This causes the protein to diffuses from the DNA thereby inducing transcription.
Usage and Biology
- Inducer: L(+)-arabinose
- L(+) - Arabinose is a sugar and is harmless. For overexpression of proteins the concentration of 0,001-0,02% Arabinose can be used.
- We designed this biobrick in order to get a promoter with low background activity so that our expressed membrane proteins wouldn’t damage the cells before induction.
- We also needed an adjustable regulation of induction so that we could induce different levels of transcription, as membrane proteins might damage the cells when expressed at high levels according to the small capacity of enrichment in the membrane.
- Can with success be combined with a promoter from pBAD SPL to obtain a very low leakiness and an appropriate strength. This is very efficient for expressing lethal proteins.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Characterization
Characterization using PET cleaving enzyme pNB13
This biobrick did measure up to our expectations as shown in the following data. We also used this promoter to express our PET cleaving enzyme pNB-Est13, which is anchored C-terminal to EstA.(E. Coli membrane anchor protein). AraC-Pbad shows a low background activity and a good respose to the induction with Arabinose. For more details: BBa_K808032
Characterization using GFP
The promoter was characterized using GFP to measure gene expression at different arabinose concentrations. Cells were grown in 24-well plates in a defined medium ([http://2012.igem.org/Team:TU_Darmstadt/Materials/GFP-Medium GFP-medium)] that provided fast growth without interfering with fluorescence measurement. Measuring in LB-medium turned out not to be possible.
Results Cells showed fluorescence after 4-5 hours when grown in a medium containing >0.01 % (w/v) of arabinose. The response of the promoter to different concentrations turned out to be dynamic, and different levels of gene expression were inducible. E. coli DH5alpha was used for the measurements.

GFP response in LB-medium
For overexpression of our genes we wanted to grow the cells in LB-medium. To find out how we had to scale and time gene expression we made another test using GFP. Because of the high background fluorescence it was required to transfer cells in H2O before measurement.
Results The response was larger in LB-medium compared to GFP-medium. Reasons for this might be the faster metabolism as well as slight presence of arabinose in LB-medium. This could lead to higher expression of receptors targeting arabinose and a faster response. Measurements were done in E. coli DH5alpha.
Characterization using GFP in S30 cell-free extract
Leakiness
To characterize the promoter's leakiness, we amplified our constructs containing BBa_K808000 in E.coli. We then performed plasmid minipreps to extract circular DNA and sequenced this DNA.
Sequencing analysis showed that we successfully amplified our part BBa_K2206006 with no fidelity errors. This composite part contains BBa_K808000 (the promoter), BBa_K2206000 (our 15b-5p toehold switch) and BBa_E0040 (GFP). Therefore BBa_K808000, the promoter, was suitable for preventing toxic levels of our part BBa_K2206006 in E. coli, demonstrating that BBa_K808000 has low levels of leakage.
However, we found some mutations in the promoter for part BBa_K2206007. This composite part contains BBa_K808000 (the promoter), BBa_K2206001 (our 27b-3p toehold switch) and BBa_E0040 (GFP). Therefore toxic levels of BBa_K2206007 were still produced. This indicates that the promoter has some leakage and may therefore be unsuitable for regulating the expression of lethal parts.
Response to varying arabinose concentrations
To characterize the promoter's response to varying arabinose concentrations, we prepared a cell free system containing BBa_K808000 and incubated it for 10 hours at 37°C. We then added 1 μl of arabinose at the concentrations of: 0.05%, 0.1%, 0.5%, 1% and 2% and measured fluorescence every 10 minutes for 10 hours at 37°C.
We found that fluorescence increased in as little as one hour and maximum fluorescence was reached after ~8 hours for all the concentrations. We found that the fluorescence increase occurred with as little as 0.05% arabinose (we did not measure lower than this) and increased with arabinose concentrations up to 0.5%. Interestingly, we saw a decline in fluorescence at 1% and 2% arabinose concentrations, but we are unsure as to why this happened.
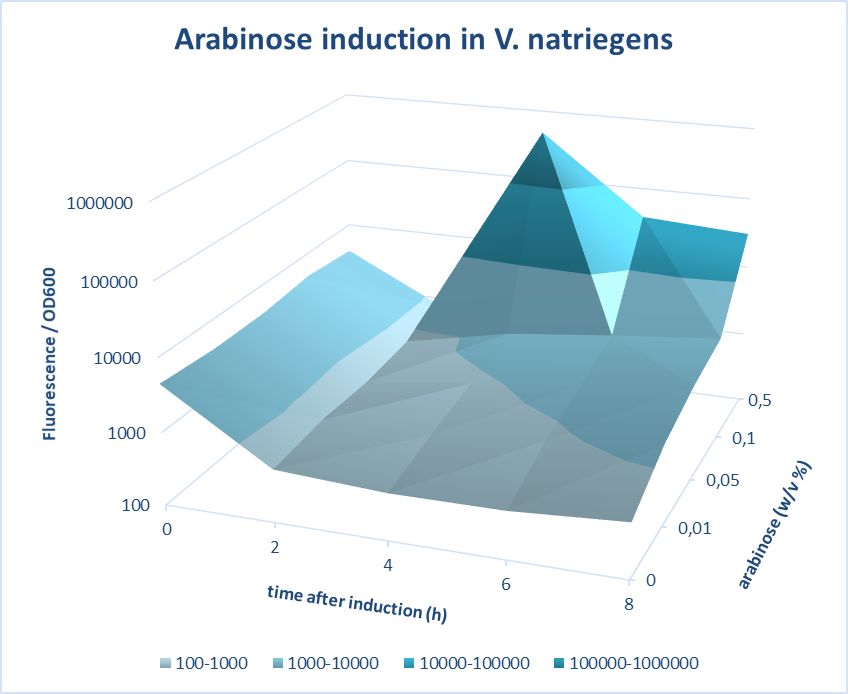
Induction of pBAD promoter in V. natriegens
(Characterized by iGEM Groningen-2019)
iGEM Groningen 2019 characterized the inducibility of pBad in V. natriegens. V. natriegens has been reported to have a remarkable doubling time of 9.8 min. As is the case for the better-studied but slower-growing E. coli, V. natriegens increases its number of ribosomes with the growth rate in order to achieve its extraordinarily high rate of protein synthesis. Multiple mechanisms contribute to this high ribosome synthesis efficiency, including high rRNA gene copy number; strong promoters that contain near-consensus −10, −35, and UP elements; and activation by the transcription factor Fis. In addition, V. natriegens rRNA promoters exhibit the relatively short-lived open-complex characteristic of rRNA promoters in E. coli, potentially contributing to the regulation of these promoters in vivo.
A construct of pBad-mCherry was designed using 3A assembly method and cloned in V. natriegens. The pBad promoter was characterized at different time points of 0, 2, 4, 6 and 8 hours after induction with different concentrations of arabinose of 0.01, 0.05, 0.1 and 0.5%. Further, the efficiency of pBad was measured by carrying out fluorescence measurements of the expression of mCherry on induction with different concentrations of arabinose. The screening was done based on quantifications of the fluorescence intensity measured. The best version of the promoter was determined based on fold change in the fluorescence intensity and low basal level. Our measurements show very high expression levels for a concentration of 0.5 % (w/v) arabinose and a very low basal level of expression without the inducer and the fold change amounts to 1127. In conclusion, pBAD offers a tight and regulation of expression in V. natriegens.
Figure 1. Characterization of Arabinose induction in V. natriegens. pBAD is a tightly regulated promoter that can be induced by arabinose in V. natriegens. Fluorescence is normalized to OD600 and presented here on a logarithmic scale. The course of fluorescence is presented for different inducer concentrations and at different time points
Reference
- Schleif, R. (2000). "Regulation of the L-arabinose operon of Escherichia coli." Trends Genet 16(12): 559-565.
- Ren, H., D. Yu, et al. (2009). "High-level production, solubilization and purification of synthetic human GPCR chemokine receptors CCR5, CCR3, CXCR4 and CX3CR1
- http://openwetware.org/wiki/Titratable_control_of_pBAD_and_lac_promoters_in_individual_E._coli_cells#pBAD_promotersOpenWetWare
- http://www.ncbi.nlm.nih.gov/pubmed/7768852?dopt=Abstract
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/positive
//classic/regulatory/other
| biology | |
| control | araC, arabinose |
| direction | Forward |
| n/a | Inducible pBad/araC promoter |
| negative_regulators | |
| o_h | |
| o_l | |
| positive_regulators | 1 |