Part:BBa_K525304
Fusion Protein of S-Layer SgsE and mCherry RFP
Fusion protein of S-layer SgsE and mCherry RFP
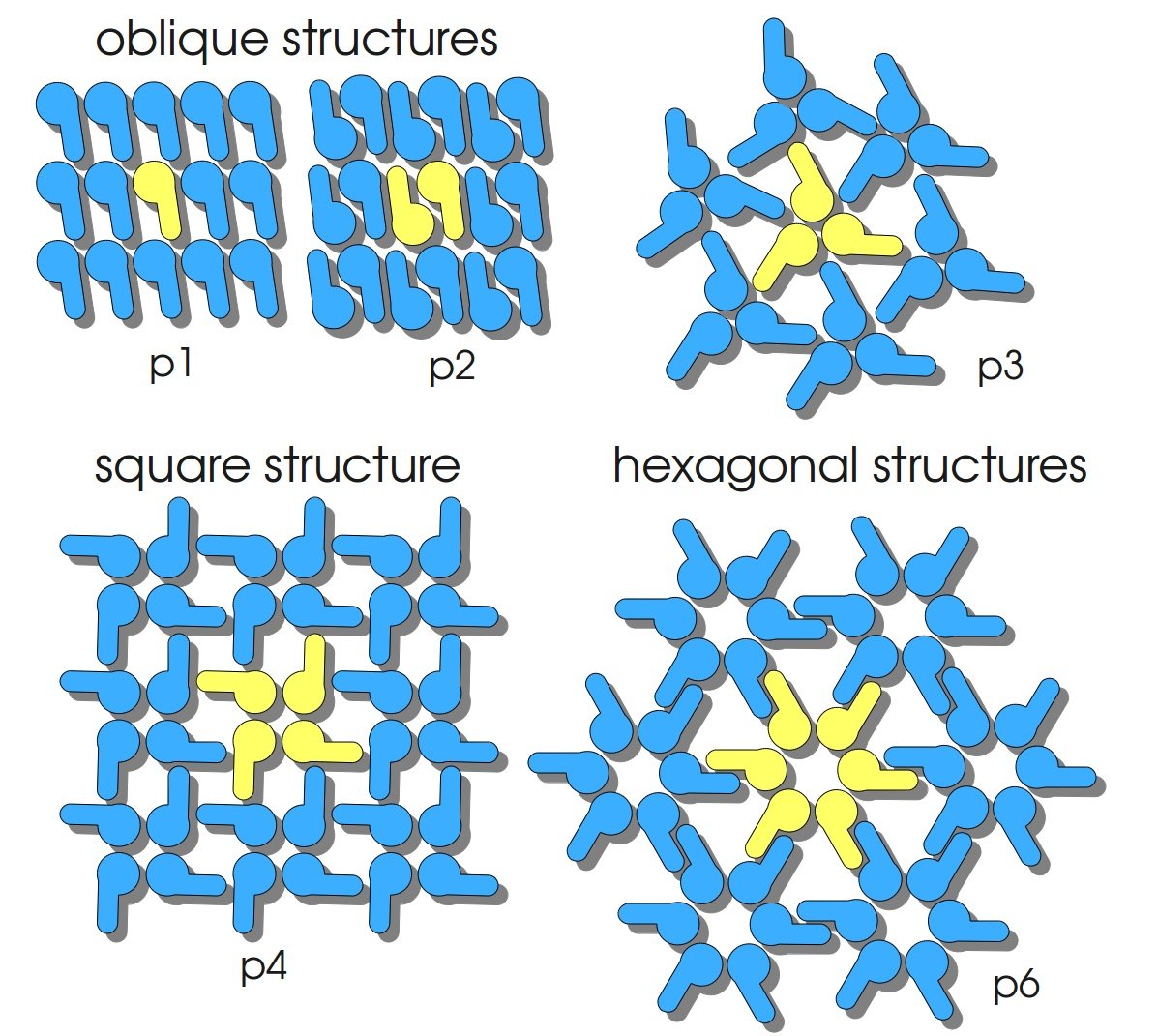
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
This fluorescent S-layer fusion protein is used to characterize purification methods and the S-layer's ability to self-assemble on surfaces. It is also possible to use the characteristic of mCherry as a pH indicator ([http://pubs.acs.org/doi/abs/10.1021/bm901071b Kainz et al., 2010]).
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | Inclusion body |
| Compatibility | E. coli KRX and BL21(DE3) | |
| Induction of expression | expression of T7 polymerase + IPTG or lactose | |
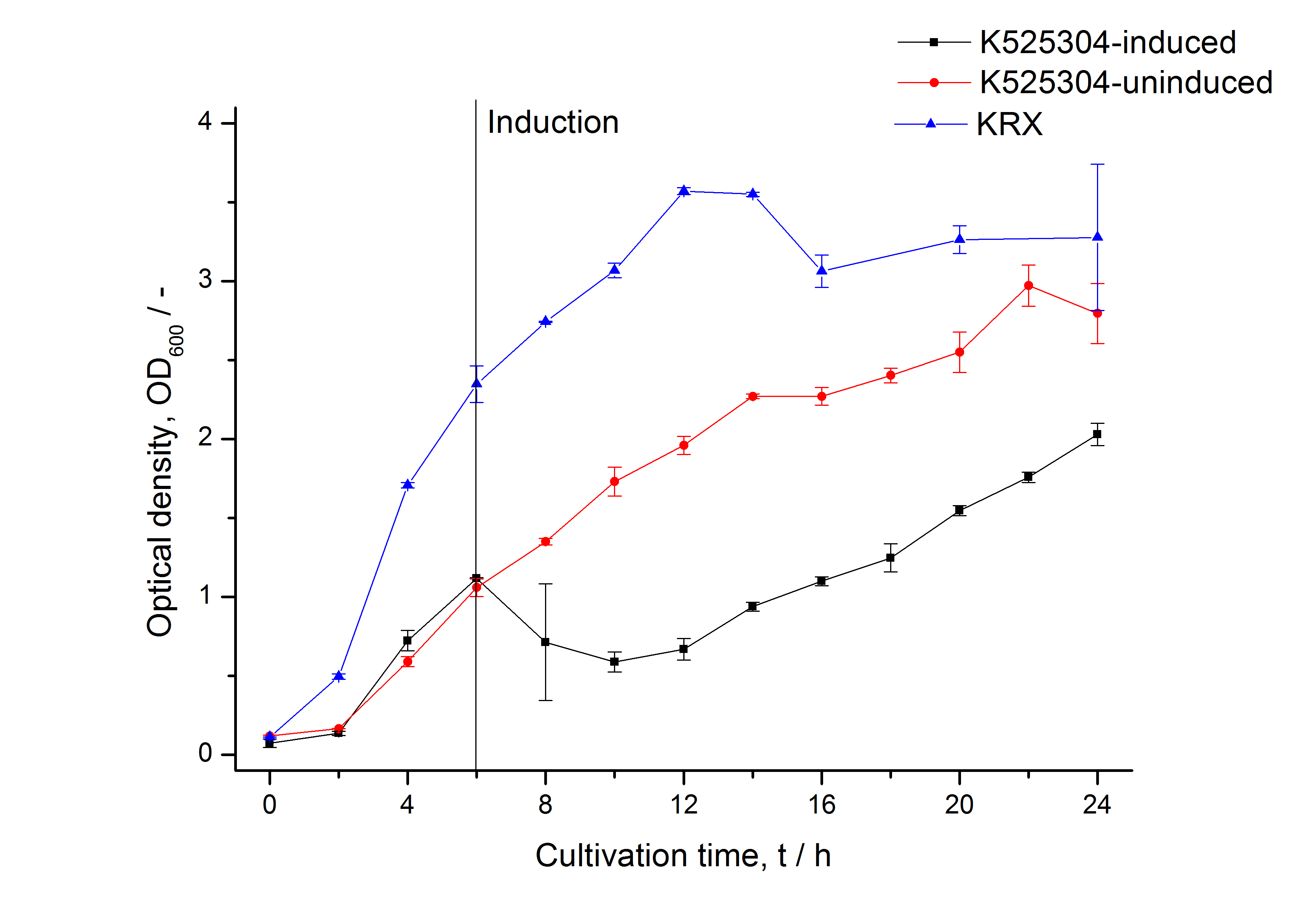
| Specific growth rate (un-/induced) | 0.128 h-1 / 0.094 h-1 | |
| Doubling time (un-/induced) | 5.41 h / 7.39 h | |
| Purification | Molecular weight | 109.9 kDa |
| Theoretical pI | 5.70 | |
| Excitation / emission | 587 / 610 nm | |
| Immobilization behaviour | Immobilization time | 4 h |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 167
Illegal BglII site found at 1022 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 76
Illegal AgeI site found at 3112 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1657
Expression in E. coli
The SgsE gene under the control of a T7 / lac promoter (BBa_K525303) was fused to mCherry (BBa_J18932) using Freiburg BioBrick assembly for characterization experiments.
The SgsE|mCherry fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0.1 % L-rhamnose and 1 mM IPTG using the autinduction protocol by Promega.

Methods
Expression of S-layer genes in E. coli
- Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX]
- Medium: LB medium supplemented with 20 mg L-1 chloramphenicol
- For autoinduction: Cultivations in LB-medium were supplemented with 0.1 % L-rhamnose and 1 mM IPTG as inducer and 0.05 % glucose
Measuring of mRFP
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 584 nm
- Emission: 620 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
References
Kainz B, Steiner K, Möller M, Pum D, Schäffer C, Sleytr UB, Toca-Herrera JL (2010) Absorption, Steady-State Fluorescence, Fluorescence Lifetime, and 2D Self-Assembly Properties of Engineered Fluorescent S-Layer Fusion Proteins of Geobacillus stearothermophilus NRS 2004/3a, [http://pubs.acs.org/doi/abs/10.1021/bm901071b Biomacromolecules 11(1):207-214].
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].
//function/reporter/fluorescence
//proteindomain/internal
| None |