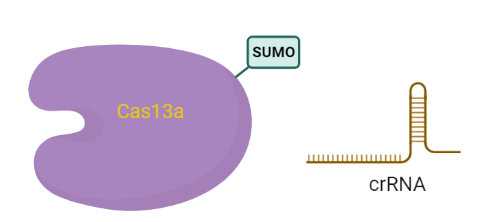
Part:BBa_K4170049
SUMO-LbuCas13a and crRNA targeting the miR-17-5P (mismatch design) coexpression system (all-in-one)
This plasmid contains all the basic genetic parts required for the simultaneous bacterial expression of codon-optimized LbuCas13a protein with the small ubiquitin-like modifier (SUMO) protein and the transcription of crRNA-5pm under the transcriptional control of T7 promoter. The aim was to develop a system where LbuCas13a and crRNA would be generated at the same time performing the CRISPR/Case13a complex directly in the bacteria. Realizing that crRNA as a delicate molecule could be degredated by ribonucleases during the storage period, we pursued the synthesis of the complex which protects the crRNA from degradations in the bacteria.
This genetic device is a part of the meta-CRISPR part collection developed by iGEM22_Thessaloniki_Meta.
Meta-CRISPR part collection
The meta-CRISPR part collection was developed in accordance with standardization, modularity and standard assembly rules aiming to enhance the CRISPR/crRNA applicability in synthetic biology projects. This collection allows the interchangeability of the genetic parts and the automation of construction following a standardized Golden Gate-based cloning strategy. Utilizing the meta-CRISPR collection, future iGEM teams can select the desired combination of DNA parts depending on the specific application of the CRISPR method (https://2022.igem.wiki/thessaloniki-meta/part-collection). Through the combination of different promoters and LbuCas13a coding sequences, one can select the desired LbuCas13a expression system, for example under the transcriptional control of the T7 or the pRha promoter. In addition, by combining the suitable genetic parts, researchers can select the purification method of the recombinant LbuCas13a protein either from inclusion bodies or from the soluble cytoplasmic fraction of the bacteria. Regarding the crRNA, utilizing the guidelines provided on the crPrep-crRNA preparation kit one can easily in silico design and produce the necessary crRNA sequence depending on the target miRNA utilizing the standardized cloning method and requiring only one additional primer. Last but not least, the LbuCas13a coding device can be assembled with the crRNA transcription system in a single plasmid enabling the simultaneous production of the LbuCas13a/crRNA complex in bacteria.

Biology and usage
According to literature references, the crRNA stability is significantly enhanced when it binds to Cas13a enzyme forming the Cas13-crRNA complex (Kick et al., 2022). The crRNAs after reconstitution with water should be stored at -80 deep freeze conditions and is prone to RNases activity. During the crRNA-related production and purification procedure, a significant amount of crRNA can be lost due to ribonuclease activity or degradation. The simultaneous coproduction of the LbuCas13a in complex with the crRNA could provide an innovative method for the development of functional CRISPR systems, enhancing the crRNA stability.
CRISPR/Cas13a system
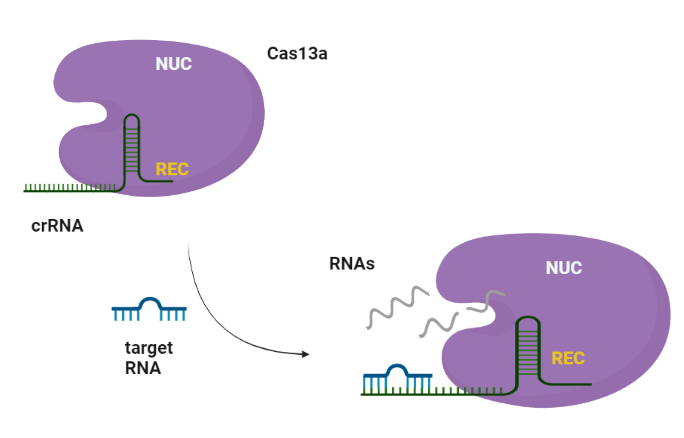
CRISPR (Clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated protein) systems are originally derived from prokaryotic adaptive immune system against invading nucleic acid components. Generally, CRISPR/Cas system can be divided into two main classes, class I and II, according to the system comprising a single or multiple effectors (Liu et al., 2017). Among them, class II (e.g., Cas9, Cas12, and Cas13) possesses more widespread application, due to its simple components (a single effector protein and a programmable guide RNA) (Wang et al., 2021). Cas13 can be further divided into four subtypes, Cas13a–d, exhibiting diverse primary sequences except the two highly conserved HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domains, which are responsible for both cis- and trans-RNase activities. (Florczuk et al., 2017) Structural studies revealed that Cas13a adopts a bilobed architecture including recognition (REC) and nuclease (NUC) lobes (Wang et al., 2021, Zhou et al., 2020.
The Cas13a enzyme guided by the specific crRNA, possesses the target-dependent trans-cleavage of all nearby RNA molecules with high recognition ability and highly processive cleavage.
Basic parts of the device construction
The part sample which is flanked at the beginning and the end with prefix and suffix respectively, is composed of the following basic parts assembled together in series and downstream of the prefix:
- Bacterial terminator (for LacI CDS): putative bacterial transcription terminator
- Biobrick Prefix sequence (backbone sequence): BioBrick prefix for parts that do not start with "ATG"
- LacI Coding sequence: Lac repressor
- LacI promoter: Promoter that has the transcriptional control of Lac repressor
- T7 promoter: promoter for bacteriophage T7 RNA polymerase T7 gene 10 (Olins and Rangwala, 1989)
- Lac operator: Lac repressor protein binding site
- Ribosome binding site: efficient ribosome binding site from bacteriophage
- 6XHis tags: 6xHis affinity tag
- LbuCas13a CDS: Cas13a protein derived from Leptotrichia Buccalis
- Thrombin site: thrombin recognition and cleavage site
- bdSUMO CDS: small ubiquitin-like modifier (SUMO) protein
- LbuCas13a CDS: Cas13a protein derived from Leptotrichia Buccalis
- T7 promoter: promoter for bacteriophage T7 RNA polymerase T7 gene 10
- crRNA-5pm CDS: coding sequence of crRNA which targets miRNA-17-5p containing a restriction site of SapI enzyme for the linearization of the plasmid
- Biobrick Suffix sequence (backbone sequence) : universal suffix for all parts
- His operon terminator (downstream of Suffix): putative transcription terminator from the E. coli
(Olins and Rangwala, 1989)
</ul>
Cloning strategy-3A Assembly
For the final construction of the plasmid we followed the 3A Assembly method which utilizes the restriction sites of the Prefix and Suffix regions to assemble the desired parts into the linearized backbone vector. Traditionally, the selection of the correct assembly products is achieved through antibiotics. Since we had all the collection of our parts into pSB1C3 backbone plasmid, we followed the guidelines of 3A assembly but the selection was based only on Chloramphenicol antibiotic.
The 3A assemply procedure is discribed in detail below:
Step 1
- Enzymatic digestion of cloned final SUMO-LbuCas13a (BBa_K4170016) plasmid with EcoRI-HF and SpeI-HF restriction enzymes.
- Enzymatic digestion of cloned crRNA-5pm (BBa_K4170049) plasmid with XbaI and PstI-HF restriction enzymes.
- Enzymatic digestion of Bba_J364007 part from the 2022 DNA distribution Kit with 'EcoRI and PstI-HF enzymes for the production of linearized pSB1C3 backbone with the desired sequences on the ends.
SUMO-LbuCas13a plasmid corresponds to the SUMO-LbuCas13a coding device under T7 promoter plasmid (BBa_K4170016 https://parts.igem.org/Part:BBa_K4170016). We selected this name as an abbreviation.
crRNA-3ps plasmid corresponds to the crRNA targeting the miR-17-5P (mismatch design) under T7 promoter plasmid (BBa_K4170049 https://parts.igem.org/Part:BBa_K4170049). We selected this name as an abbreviation.

Step 2
The digested parts were ligated by T4 ligation for the efficient construction of Cloned LbuCas13a-SUMO with crRNA-3ps plasmid. The products of the 3A assembly underwent transformation into E.coli DH5a competent cells. To verify the successful 3A assembly, colony PCR was performed using the VR and VF2 primers. Then, the samples were loaded and run in 1% agarose gel electrophoresis. As depicted on the follwoing figure from all colonies the desired SUMO-LbuCas13a-crRNA part has been amplified. For the colony PCR procedure, half amount of each colony was picked from the agar plate and diluted in 10μl of dH2O. 1μl of the H2O with the colony was used as a template DNA of the colony PCR. The other half amount was picked for the overnight liquid culture.

After applying the 3A assembly method to create the cloned SUMO-LbuCas13a-crRNA-17-5pm plasmid, we observed that our experiment was crowned with success. In our experiment the upstream part was Cas13a (5422bp) and the downstream part was the crRNA-17-5pm molecule (approximately 100bp). The destination vector was the pSB1C3 backbone vector (2029bp). According to the 3A assembly the vector must have a different antibiotic resistance than the two input plasmids. However, in our case, the vector had the same antibiotic as the input parts but the result was still successful. Thus, we can safely support that 3A assembly is an effective method for the integration of inserts with high difference in size.
SUMO-LbuCas13a-crRNA-17-5pm plasmid corresponds to SUMO-LbuCas13a and crRNA targeting the miR-17-5P (mismatch design) coexpression system (all-in-one).We selected this name as an abbreviation. Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1453
Illegal BglII site found at 5047 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1421
Illegal AgeI site found at 1910
Illegal AgeI site found at 3125 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 5476
Citations
[1] Butt, T., Edavettal, S., Hall, J. and Mattern, M., (2005) "SUMO fusion technology for difficult-to-express proteins." Protein Expression and Purification, 43(1), pp.1-9.
[2] Florczuk, M., Szpechcinski, A., & Chorostowska-Wynimko, J., (2017) "miRNAs as Biomarkers and Therapeutic Targets in Non-Small Cell Lung Cancer: Current Perspectives." Targeted Oncology, 12(2), 179-200.
[3] Garner, K., (2021) "Principles of synthetic biology." Essays in Biochemistry, 65(5), pp.791-811.
[4] Kick, L., von Wrisberg, M., Runtsch, L. and Schneider, S., (2022) "Structure and mechanism of the RNA dependent RNase Cas13a from Rhodobacter capsulatus." Communications Biology, 5(1).
[5] Liu, L., Li, X., Ma, J., Li, Z., You, L., Wang, J., Wang, M., Zhang, X.and Wang, Y., (2017) "The Molecular Architecture for RNA - Guided RNA Cleavage by Cas13a." Cell, 170(4), pp .714 - 726.e10.2.
[6] Wang, B., Zhang, T., Yin, J., Yu, Y., Xu, W., Ding, J., Patel, D. and Yang, H., (2021) "Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems." Molecular Cell, 81(5), pp.1100-1115.e5
| None |