Part:BBa_K3453079
Grapevine VvTrnL-UAA Toehold Switch 3.1
This part is a toehold switch sensor for sequence-based detection of grapevine. It targets a fragment of the TrnL-UAA gene of Vitis vinifera (BBa_K3453083).
Together with BBa_K3453080, this part is a functional improvement of BBa_K2916050, BBa_K2916051, BBa_K2916052, BBa_K2916053 and BBa_K2916054.
Usage and Biology
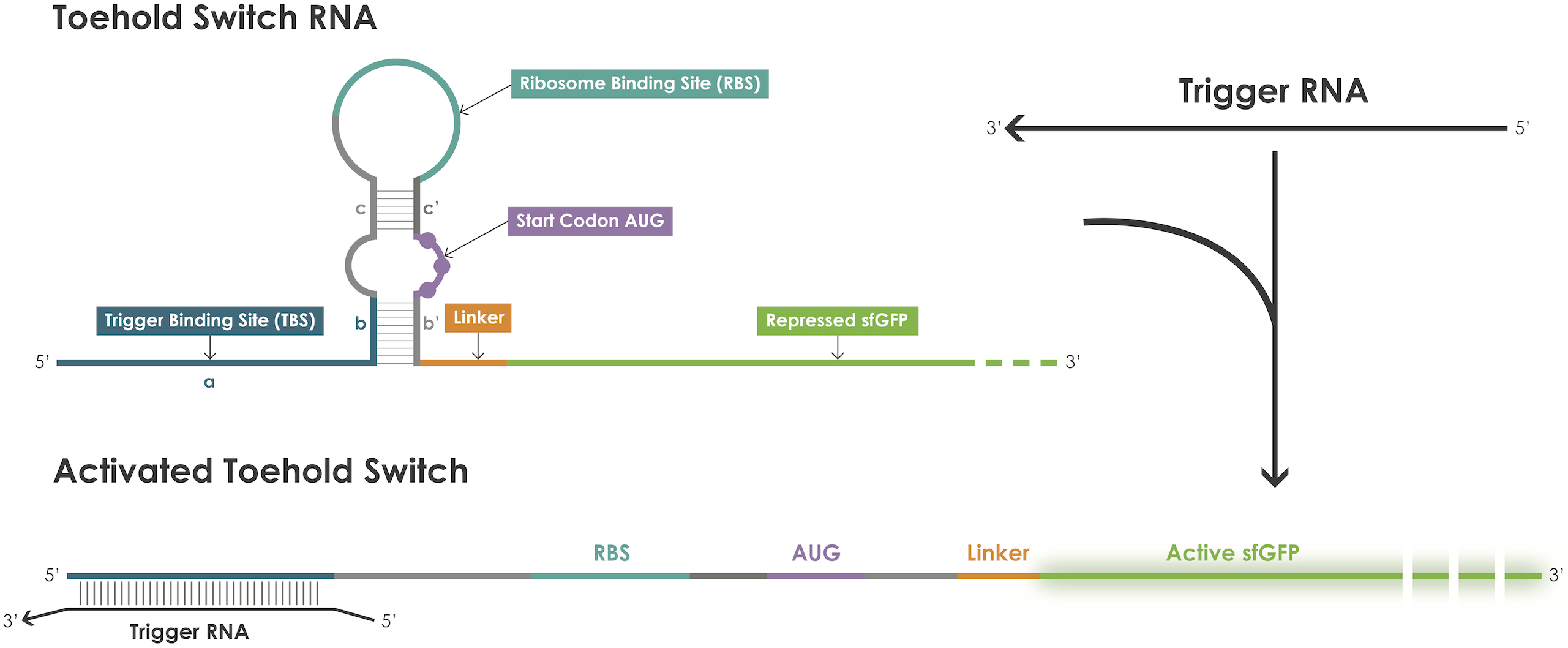
A toehold switch is an RNA–based device containing a ribosome binding site (RBS) and an ATG start codon embedded in the middle of a hairpin structure that blocks translation initiation [1]. The hairpin can be unfolded upon binding of a trigger RNA thereby exposing the RBS and the ATG start codon and thus permitting translation of the reporter protein (Figure 1).
Figure 1. Toehold switches principle.
This part is a toehold switch sensor that targets a fragment of the TrnL-UAA gene of Vitis vinifera (BBa_K3453083). It was designed using the web tool developed by To et al. [2] and follows the architecture of the Series B of toehold switch sensors for Zika virus detection [3] and of the BioBits™ toeholds [4]. It's secondary structure predicted by NUPACK web server [5] using default parameters is represented in Figure 2 and here-after in dot-bracket notation.
........(((.....))).(((((((((..(((((((.(.(((((............))))).).)))))))..)))))))))......(((....))).....
Figure 2. Secondary-structure prediction of this part with the ATG of the reporter gene. The prediction was realised using the NUPACK web server [5] with default parameters and graphically represented using the forna RNA secondary structure visualization tool [6]. Nucleotides were coloured to match the different segments in Figure 1.
This part is a functional improvement of BBa_K2916050, BBa_K2916051, BBa_K2916052, BBa_K2916053 and BBa_K2916054, five grapevine toehold switches designed by the EPFL 2019 team, all having the trigger binding site in the same orientation as in the transcriptional orientation of their target gene (TrnL-UAA gene of grapevine) and of the trigger sequences derived from (BBa_K3453081 and BBa_K3453083). These five grapevine toehold switches will most likely not be able to detect the presence of an RNA naturally expressed in grapevine because the trigger binding site is not in a compatible orientation. Indeed, as highlighted in Figure 1, the binding between the trigger and the switch is dictated by RNA-RNA interactions between the complementary base pairs. As a result, this trigger / switch pair does not interact and is not functional.
However, the hairpin structure of the grapevine switch 1.1 is predicted to be unfolded by the reverse complements of the two triggers BBa_K3453082 and BBa_K3453084 (Table 1).
To improve the functions of these 5 grapevine toehold switches designed by the EPFL 2019 team, we designed 2 new toehold switches that we’ll refer to as Grapevine Switch 3.1 (this part BBa_K3453079) and Grapevine Switch 3.2 (BBa_K3453080) having the trigger binding sites in the correct orientation and showed experimentally that they are able to respond to an RNA molecule having the transcription orientation of the grapevine TrnL-UAA gene (BBa_K3453081 and BBa_K3453083) (Table 1).
| Table 1. Grapevine triggers. | |||
|---|---|---|---|
| Label | Part number | Transcriptional units’ part numbers | Source |
| Grapevine trigger 'short' | BBa_K3453081 | BBa_K3453181 | EPFL 2019 iGEM team personal communication |
| Grapevine trigger 'short reverse' | BBa_K3453082 | BBa_K3453182 | Reverse complement of BBa_K3453081 |
| Grapevine trigger 'long' | BBa_K3453083 | BBa_K3453183 | designed by the EPFL 2019 iGEM team and its sequence was deduced from “Amplification” page of the EPFL 2019 wiki where is described how the triggers were designed: the grapevine trigger was PCR amplified using as template the Vitis vinifera TrnL-UAA and the forward primers EC_fwd1 or EC_fwd1_T7 and the EC_rev1 reverse primer (it should be noted that the sequences of all reverse primers listed in the additional materials of this page are given as 3’-5’ sequences and not in the conventional 5’-3’ orientation). |
| Grapevine trigger 'long reverse' | BBa_K3453084 | BBa_K3453184 | Reverse complement of BBa_K3453083 |
The functionality of this part was tested using sfGFP-LVAtag (BBa_K2675006) as a reporter. The expression was controlled by the T7 promoter (BBa_K2150031) and the strong SBa_000587 synthetic terminator (BBa_K3453000) in the composite part BBa_K3453179.
For toehold switch characterisation, the BBa_K3453179 was assembled in the low copy plasmid pSB3T5. The trigger sequences (Table 1) were placed under the control of the T7 promoter and followed by the strong SBa_000587 synthetic terminator for the T7 RNA polymerase and the resulting transcriptional units were synthesized and assembled in the high copy plasmid pSB1C3. The sensor and the trigger plasmids were both introduced in E. coli BL21 Star™(DE3) cells (Thermo Fisher Scientific), genotype F- ompT hsdSB (rB-mB-) gal dcm rne131 (DE3). As all BL21(DE3) E. coli strains, these cells contain the T7 RNA polymerase under control of the lacUV5 promoter and thus require IPTG to induce expression. The particularity of BL21 Star™(DE3) cells is that they contain a truncated version of the RNaseE gene (rne131) that leads to reduced level of mRNA degradation and thus increased RNA stability. For fluorescence measurements, E. coli cells containing switch and trigger plasmids were first grown overnight in 96-deep-well plates containing 1 mL of LB medium supplemented with 5 µg/mL tetracycline and 17.5 µg/mL chloramphenicol, then diluted by 40x into similar media. Upon reaching early log-phase, cells were further diluted 20x in 100 µL of LB medium supplemented with 5 µg/mL tetracycline, 17.5 µg/mL chloramphenicol and 10 µM IPTG in an opaque wall 96-well polystyrene microplate, the COSTAR 96 (Corning). The plate was incubated at 37°C at 200 rpm and the sfGFP fluorescence (λexcitation 483 nm and λemission 530 nm) and OD600nm were measured every 10 min for 24 hours in a CLARIOstar (BMGLabtech) plate reader. Fluorescence values were normalised by OD600nm and, using the calibration curves presented on the ‘Measurement’ page of our wiki, we converted the arbitrary units into Molecules of Equivalent FLuorescein (MEFL) / particle.
The results presented in figures 3, 4 and 5 show that this part, the Grapevine Switches 3.1 is functional and behave as expected.
The sfGFP expression from Grapevine Switch 3.1 was detected only in the presence of the Grapevine triggers (both short and long) when transcribed in the natural direct orientation. As control, this Grapevine Switches 3.1 was also challenged with the reverse triggers and, as expected, no sfGFP expression was detected. Moreover, sfGFP expression from Grapevine Switch 3.1 in BBa_K3453179 was high and at a level comparable to that of the two positive controls (BBa_K3453104 and BBa_K3453105) which are equipped with two strong RBS (BBa_K2675017 and BBa_K3453005) with comparable strength. The results presented in figure 3 show also that, as expected, no fluorescence was detected in the negative controls as either the sfGFP gene was not present or the promoter and the RBS were absent.
Figure 3. In vivo characterization of sfGFP expression by E. coli BL21 Star™(DE3) cells harbouring the grapevine toehold switch 3.1 and the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative controls have been performed with an empty pSB3T5, pSB1C3 (no trigger) and BBa_K3453103 (no promoter, no RBS) and the positive controls with BBa_K3453104 and BBa_K3453105. The data and error bars are the mean and standard deviation of at least three measurements on independent biological replicates.
Figure 4. MEFL / Particle fold changes of the grapevine toehold switch 3.1 in the presence of the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184) compared to the MEFL / Particle value in the absence of the trigger.
Figure 5: Pictures of E. coli BL21 Star™(DE3) cells harbouring the expression cassettes of sfGFP-LVAtag under the control of the grapevine toehold switch 3.1 and in the presence of the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative control was performed with an empty pSB1C3 (no trigger).
In contrast, the Grapevine Switch 1.1 (BBa_K2916050) is not functional in the same experimental setup (BBa_K3453171): no sfGFP expression was detected in the presence of the Grapevine triggers (both short and long) when transcribed in the natural direct orientation (Figure 6). As in this part, the trigger binding site is in the opposite direction, it is able to respond to the presence of a trigger artificially expressed in E. coli in reverse orientation.
Figure 6. In vivo characterization of sfGFP-LVAtag expression by E. coli BL21 Star™(DE3) cells harbouring the grapevine toehold switches 1.1 (BBa_K2916050) and 3.1 (this part) and the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative controls have been performed with an empty pSB3T5, pSB1C3 (no trigger) and BBa_K3453103 (no promoter, no RBS) and the positive controls with BBa_K3453104 and BBa_K3453105. The data and error bars are the mean and standard deviation of at least three measurements on independent biological replicates.
Conclusions
We have successfully built this parts, the Grapevine Switch 3.1 (BBa_K3453079), which is able to act as a toehold switch and (i) efficiently repress the downstream reporter gene expression in the absence of the cognate trigger and (ii) release the translation inhibition in the presence of the cognate trigger. Moreover, the cognate trigger is an RNA molecule having the transcription orientation of the TrnL-UAA gene of grapevine (BBa_K3453181 and BBa_K3453183).
Thus, we have made a functional improvement for 5 grapevine toehold switches designed by a previous iGEM team (BBa_K2916050, BBa_K2916051, BBa_K2916052, BBa_K2916053 and BBa_K2916054) that do not have the trigger binding sites in the correct orientation and that are not able to detect the presence of an RNA naturally expressed in grapevine as our Grapevine Switch 3.1 (this part BBa_K3453079) does.
References
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939.
[2] To AC, Chu DH, Wang AR, Li FC, Chiu AW, Gao DY, Choi CHJ, Kong SK, Chan TF, Chan KM, Yip KY. A comprehensive web tool for toehold switch design. Bioinformatics (2018) 34, 2862-2864.
[3] Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell (2016) 165, 1255-1266.
[4] Huang A, Nguyen PQ, Stark JC, Takahashi MK, Donghia N, Ferrante T, Dy AJ, Hsu KJ, Dubner RS, Pardee K, Jewett MC, Collins JJ. BioBits™ Explorer: A modular synthetic biology education kit. Sci Adv (2018) 4, eaat5105.
[5] Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. Journal of Computational Chemistry (2011) 32, 170–173.
[6] Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics (Oxford, England) (2015) 31, 3377–3379.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |