Part:BBa_K3147008
I. Part BBa_K3147008 function
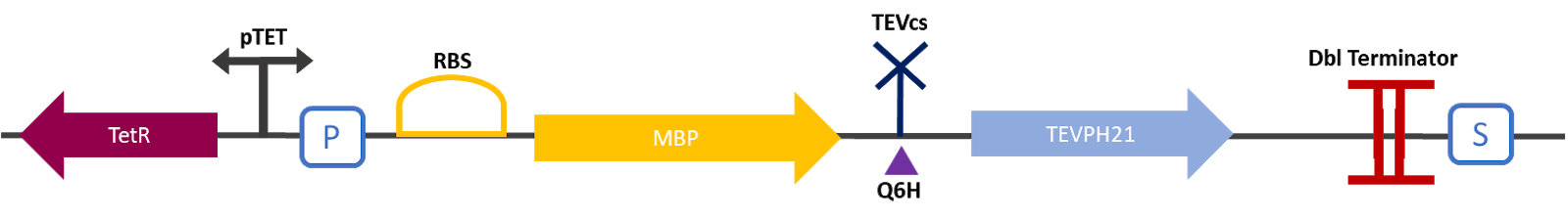
The 2019 Montpellier iGEM team made this construction in order to be able to compare the basal activity of the TEV protease (Part:BBa_ K1319004) with the mutant TEV protease TEVPE21 [3]. A modified TEV cut-off site was added between the MBP and the protease. MBP increases the solubility of the fusion protein [1], preventing the aggregation of the protein of interest. This stabilizes the expression and the sequence of the produced MBP does not have a signal peptide, which allows the protein to remain in the cytosol. The TEV cutting site allows self-cleavage of MBP from TEV once the protein is produced [2].
II. Proof of function
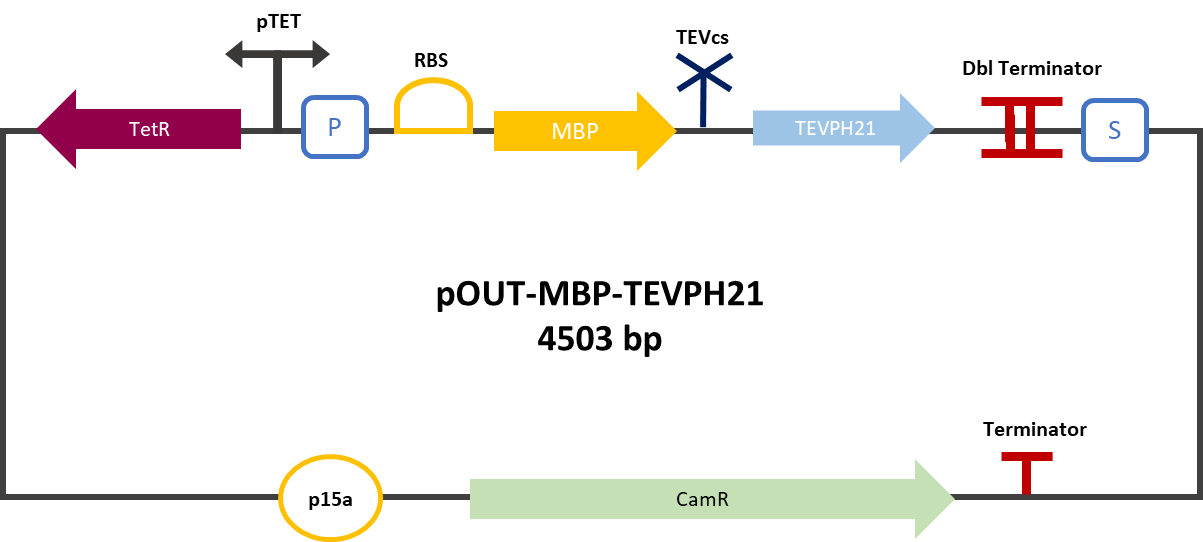
The experimental approach that has been used to test the protease activity is to compare the fluorescence restoration rate of sfGFP compared to MBP-TEV and MBP-TEVPEH21. The construction was cloned by Gibson Assembly in a pOUT18 backbone under the control of a TET ON promoter, in order to control its expression. The TEV cutting site used is the mutated site: ENLYFH/G this mutation allows theoretically the mutant TEV to better cleave this sequence according to this source [3].
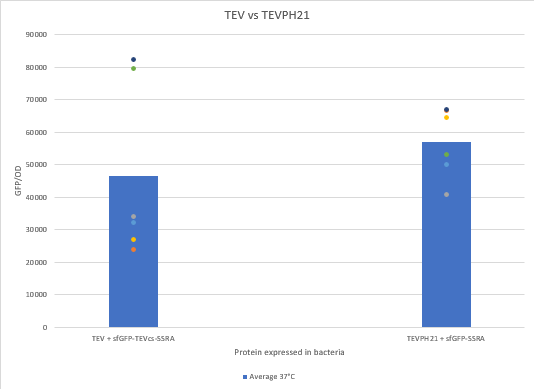
In this experiment, basal controls of maximum and minimum fluorescence of the reporter genes were used. Fluorescence data are normalized by fluorescence divided by optical density. To carry out these experiments, E. coli from the NEB10β strain were co-transformed using both constructions. The protease is expressed by inducing the TET promoter with 50 ng/mL of aTc (anhydrotetracycline).
We can see that the TEVPH21 is more efficient than the TEV WT. We think it’s because our TEV WT isn’t the one in the article where we found this mutant.
Reference
[1] Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589.
[2] Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41.
[3] Yi, L. et al. 2013. « Engineering of TEV Protease Variants by Yeast ER Sequestration Screening (YESS) of Combinatorial Libraries ». Proceedings of the National Academy of Sciences 110(18): 7229‑34 Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 77
Illegal SapI.rc site found at 1868
| None |