Part:BBa_K3110042
lldRO1-J23117-lldRO2 Weak RBS sfGFP
This part contains the lldRO1-J23117-lldRO2 attached to a weak RBS, sfGFP (Reporter Protein) and double terminator.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 78
Illegal NheI site found at 101 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 173
Usage and Biology
The regulatory element lldR binds to the lldRO1 and lldRO2 (the operator regions) and inhibit transcription. J23117 is a promoter intercalated between the operators. lldR dimer represses the transcription possibly by forming a DNA loop which doesn’t allow the RNA polymerase to bind the promoter. Upon binding of L-Lactate to LldR, however, this transcriptional suppression is lost and instead lldR complex with L-Lactate remains bound to LldRO1 acting as a transcriptional activator.
Characterization of the promoter
We wanted to recheck how this part would respond to different lactate concentration. To efficiently produce our reporter protein (sfGFP), that will later be replaced by YebF-IL12, we have used RBS of different strengths. Part:BBa_K3110040, Part:BBa_K3110041, Part:BBa_K3110042
Genetic design for characterization with LldR
To test this part we have designed two plasmids:
- O1-P-O2 -weak RBS-sfGFP-terminator in a medium copy plasmid pSB1C3
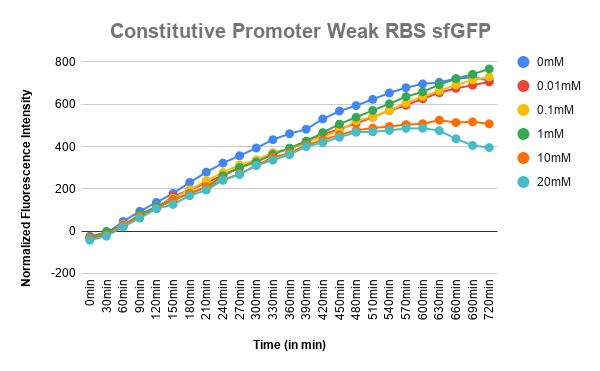
- P-weak RBS-sfGFP-terminator in another pSB1C3 (Same Promoter without the operator regions) - as a control
So, in the presence of lactate, L-Lactate should bind to LldR and enhance the expression of sfGFP where we have O1PO2 upstream of sfGfP.
Experimental Procedure
- We chose BL21(DE3) as our chassis and transformed both the plasmids in BL21(DE3) separately. Cultured them in M9-Glycerol(with Chloramphenicol of concentration 34 ng/ul) for 14-16 hrs at 37C, 200 r.p.m.(Primary Culture). After that, we put the secondary culture of both at an OD of O.1 in M9-Glycerol medium (With Chloramphenicol concentration- 34 ng/ul) and kept at 37C for 5-6 hours.
- Once the OD of the secondary culture reached 0.4, we transferred the culture to 96 well microtiterplate with appropriate blank (M9 + required lactate concentration) after induction with different concentrations of lactate (0 mM, 0.01 mM, 0.03 mM, 0.06 mM, 0.1 mM, 0.3 mM, 0.6 mM, 1 mM, 3 mM, 6 mM, 10 mM, 13 mM, 16 mM, 20 mM) and kept in plate reader. Our assays were done in technical triplicates and repeated thrice with different biological samples.
- We used the Gen5 software to program the plate reader. The cultures were kept at 37degC with orbital shaking at 282cpm for 27:30 mins. This was followed by reading OD (at 600 nm) and then fluorescence at 480 excitation and 510 emission with 70 gain. This was iterated over cycles set for 8hrs.
- Analysed the data after completion of the program.
Results
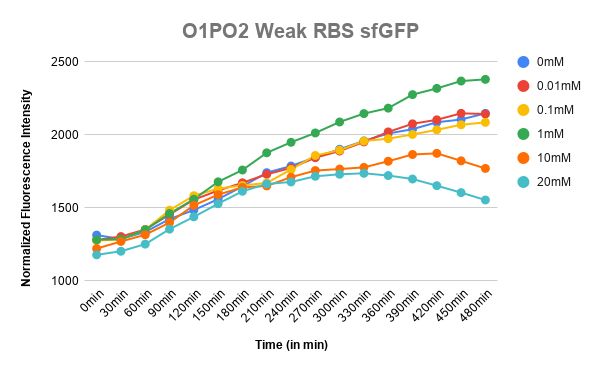
Corrected Fluorescence = Fluorescence (Sample) - Fluorescence (Blank) Corrected OD = OD (Sample) - OD (Blank) Normalized Fluorescence = Corrected Fluorescence / Corrected OD Plotted the graph of Normalized Fluorescence with time and also against different lactate concentrations at 480 min time point.
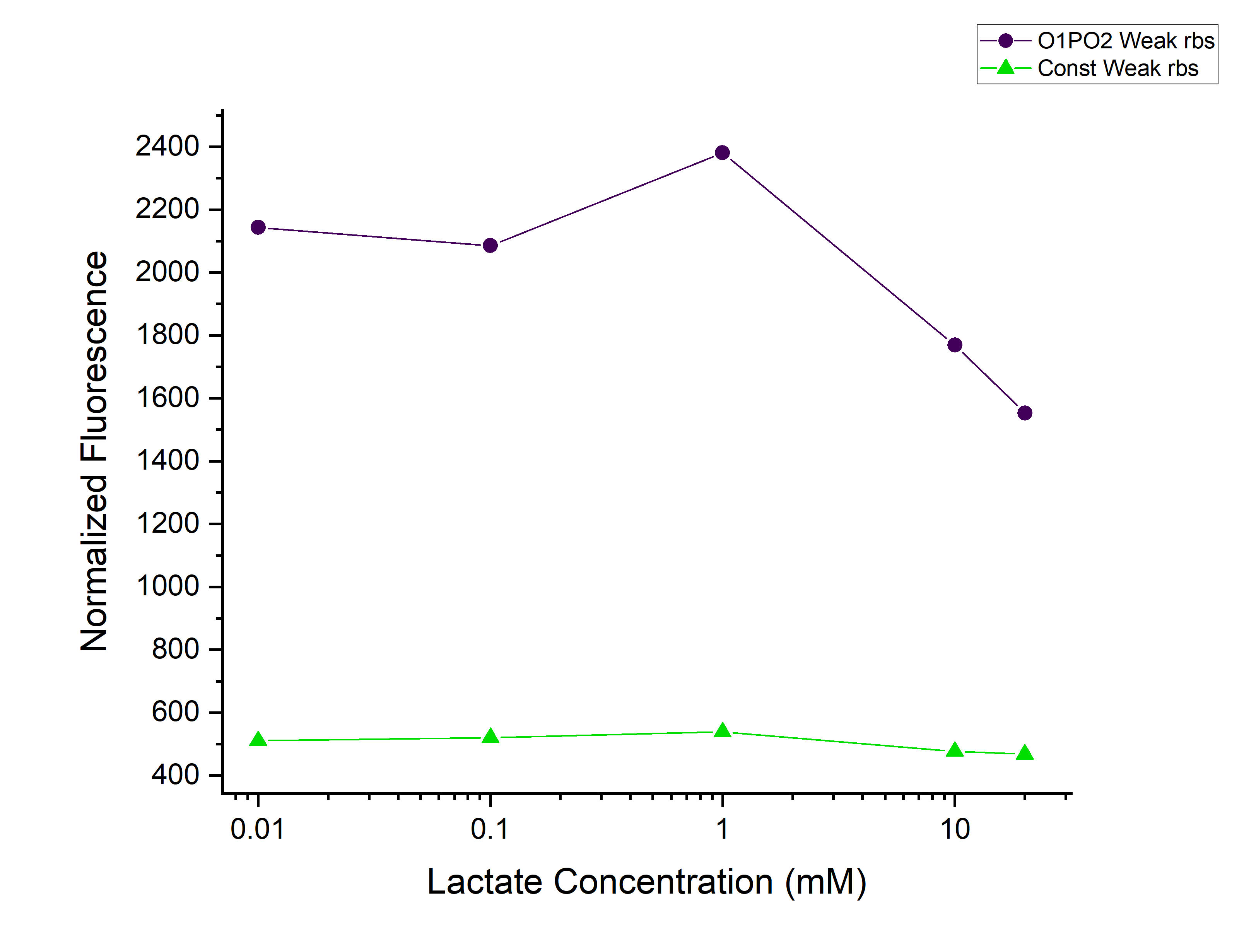
As can be seen in the graph, when P-RBS-sfGFP is there, the fluorescence level is not varying much with an increase in L-lactate concentration. But, in the case of O1PO2-RBS-sfGFP, fluorescence level has increased significantly. The absence of operator sites in the control sequence leads to no response of the construct towards changing concentrations of lactate. However, in our test sample, in the presence of lactate, lactate binds to LldR and acts as a transcriptional activator leading to a sigmoidal expression of sfGFP. As the lactate concentration is increasing, the amount of free LldR(Repressor) inside the cell is decreasing as well as the LldR bound to Lactate (activator) is increasing - so, the rate of sfGFP expression is also increasing with higher Lactate (up to a certain concentration). After lldR is saturated with L-Lactate, the sfGFP expression becomes steady. The lactate concentration of around 20mM seems to be toxic to the bacteria, hence causing slightly lower sfGFP expression. This is contradictory to the literature which mentions lactate concentration than 100mM is toxic and hence further validation for this point is required.
| None |