Part:BBa_K292006:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K292006
Reliable to use when we want to control a gene expression by using pLac promoter. In fact when LacI is not expressed, pLac is activated, and when there is an expression of LacI, pLac is inhibited. We can easily design a system to induce the transcription of LacI by using another operon as the tetracycline operon and creates a failed feedback mechanism for pLac inhibition and so, for gene expression inhibition.
User Reviews
UNIQc5f0edf2c3d62d7f-partinfo-00000000-QINU
|
•
NTNU_Trondheim 2011 |
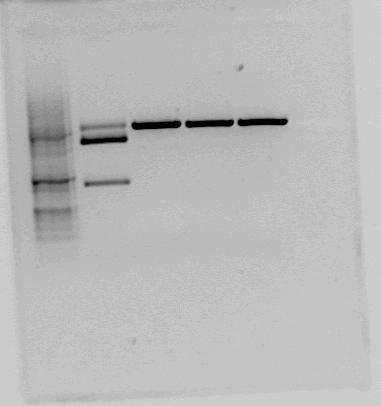
We found and orderd this part from the parts registry. Since it had RBS and double terminator it would save us from some cloning work. When testcutting the constructs containing BBa_K292006 we got some unexpected results. Therefore we decided to digest the brick with three differnt enzymes with restriction sites in the given biobrick-sequence in addition to EcoR1 and PstI (for looking at the biobrick length) Except from the sample cut with EcorR1 and PstI, the samples were also cut with BglI wich has a recognition site in the pSB1A2 backbone. Overwiev of restrctions and expected fragmentlengths:
As we can see on the resulting gel none of the restriction sites inn the LacI2 fragment appear to be present. In additon the LacI2 insert looks smaller than it's supposed to be. This indicates that the biobrick sequence does not match the sequence given in the parts registry.
|
UNIQc5f0edf2c3d62d7f-partinfo-00000002-QINU
UNIQc5f0edf2c3d62d7f-partinfo-00000003-QINU
|
••
NTNU_Trondheim 2012 |
We decided to improve this part since last years team had problems with it. We ordered the biobrick from the registry, test cut it, and sequenced it. In addition we reassembled the the parts BBa_B0030, BBa_C0012 and BBa_B0014, test cut these, and sent them to sequencing. Both BBa_K822004 and BBa_K292006 was also investigated using gel electrophoresis. The gel pictures are given below:
When the cloning work was done, we sent both our new biobrick and the old one (BBa_K292006) to sequencing. The sequencing results can be found [http://2012.igem.org/Team:NTNU_Trondheim/Sequencing_Improved_Construct here]. The sequencing result shows that in the old biobrick, only the terminator is present, and no LacI or RBS. In our improved biobrick, both RBS, LacI and terminator are present. |
|};
UNIQc5f0edf2c3d62d7f-partinfo-00000008-QINU