Part:BBa_K1761006
SmallBit Split Luciferase
SmallBit is the small part of a Split Luciferase. Its molecular weight is 1 kDa. This construct on its own has no function since SmallBit on itself has no luminescence activity. For luminescence activity, the LargeBit of the Split Luciferace is needed.
Sequence has been published by iGEM TU-Eindhoven 2017.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Split luciferases can be used as signaling components. This split luciferase consists of two parts, namely LargeBit and SmallBit. These parts were both inserted in a pETDuet-1 vector together with OmpX and a linker. LargeBit and SmallBit have an affinity towards each other, so when in close proximity they will come together and will give a luminescence signal. Followed is a short description of each part and of the NanoBit construct.
OmpX - SmallBit BBa_K1761006 [1]
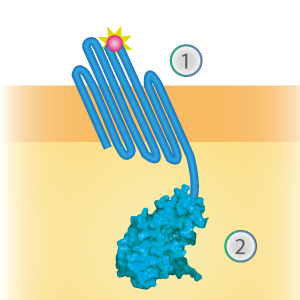
SmallBit is the small part of this Split Luciferase. Its molecular weight is 1 kDa, it is only 11 amino acids long. OmpX (1) (with a correct mutation for the amber stop codon TAG), a BsoBI-linker and SmallBit (2) together will look like Figure 1. This construct on its own has no function since SmallBit on itself has no luminescence activity.
Figure 1: Schematical overview of the OmpX - SmallBit construct.
OmpX - LargeBit BBa_K1761005
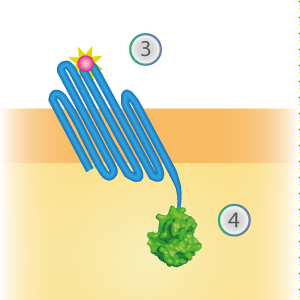
LargeBit is the big part of this Split Luciferase. Its molecular weight is 18 kDa. OmpX (1) (with a correct mutation for the amber stop codon TAG), a BamHI-linker and LargeBit (2) together will look like Figure 2. This construct on its own has no function since LargeBit on itself has no luminescence activity.
Figure 2: Schematical overview of the OmpX - LargeBit construct.
NanoBit construct
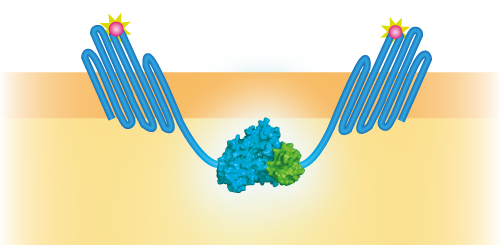
NanoBit utilizes a structural complementation-based approach to monitor protein interactions within living cells. Protein interaction promotes structural complementation and generation of a bright, luminescent enzyme. Protein dynamics can be followed in real-time in living cells following addition of the Nano-Glo Live Cell Reagent, a non-lytic detection reagent containing the cell-permeable furamizine substrate. See Figure 3 for the whole construct.
Figure 3: Schematical overview of the NanoBit construct.
Characterization
When mutated with the amber stop codon TAG, a unnatural amino acid with an azide-functionalized group can be expressed. After the expression of this amino acid, OmpX can covalently bind almost anything, as long as it contains a DBCO-functionalized group. The binding finds place by using a bio-orthogonal “click” reaction (SPAAC chemistry). To test the functionality of this “click” reaction, some experiments were done by clicking DBCO-PEG4-TAMRA at the surface. For all the experiments, the following vectors were used: pETDuet-1 with one or two construct(s) inserted (OmpX + intracellular protein) and pEVOL-pAzF (tRNA + tRNA synthetase, see BBa_K1492002 [2]). Both vectors were transformed into BL21(DE3). The expression was introduced by adding arabinose, IPTG and the unnatural amino acid.
DBCO-PEG4-TAMRA Confirmation
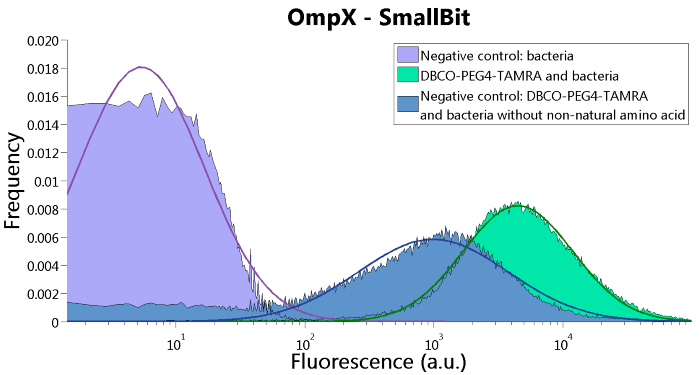
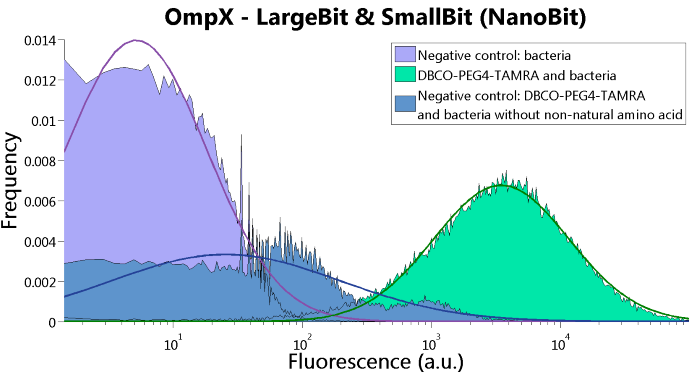
To confirm whether OmpX is in the membrane and whether or not the unnatural amino acid is being incorporated into OmpX, DBCO-PEG4-TAMRA was used. TAMRA is a fluorescent dye that can be used to verify the “click” reaction. If the unnatural amino acid is present, DBCO-PEG4-TAMRA should “click” to the transmembrane protein OmpX and stay there. This can be analyzed with FACS. For more information about how to perform FACS experiments, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols].
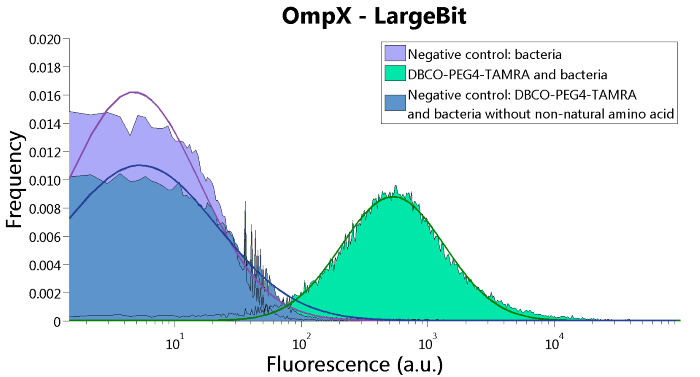
To verify that OmpX is in the membrane, we used the OmpX – LgBiT and OmpX – SmBiT constructs. These gave the following results after clicking with DBCO-PEG4-TAMRA (see Figure 4, 5 and 6). From this it can be concluded that OmpX is in the membrane and that the “click” reaction works.
Figure 4: FACS results of OmpX - LargeBit.
Figure 5: FACS results of OmpX - SmallBit
Figure 6: FACS results of OmpX - LargeBit and OmpX - SmallBit.
Bioluminescence Confirmation
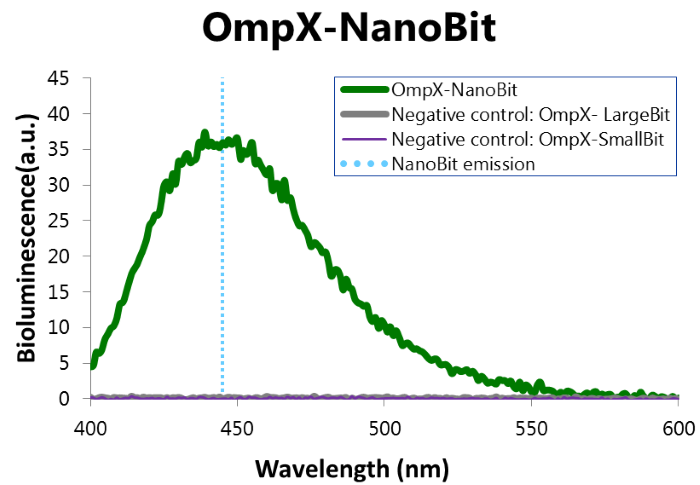
To confirm whether NanoBit is present and is working, a bioluminescence measurement was performed. The results of this experiment are shown in Figure 7.
Figure 7: Bioluminescence of results of the NanoBit construct.
Characterization iGEM 2016
Besides linking SmallBiT and LargeBiT to OmpX, it is also possible to use the NanoBiT system in other systems. iGEM TU Eindhoven 2016 (http://2016.igem.org/Team:TU-Eindhoven) used the NanoBiT system to do in vitro assays on T14-3-3 scaffold proteins. SmallBiT and LargeBiT were linked to CT52, a protein which has high affinity for the T14-3-3 scaffold if the small molecule fusicoccin is present.
CT52-SmallBiT: "https://parts.igem.org/Part:BBa_K2065000"
CT52LargeBiT: "https://parts.igem.org/Part:BBa_K2065007"
This system was used as a read out method for T14-3-3 heterodimers. Under influence of a small molecule, SmallBit and LargeBit attached to CT52 dimerize on the scaffold, leading to activation of NanoLuc Luciferase and emission of bioluminescence light. It is schematically visualised in figure 1. For more detailed information about the application of CT52-SmallBiT visit our wiki "http://2016.igem.org/Team:TU-Eindhoven/Read-out".
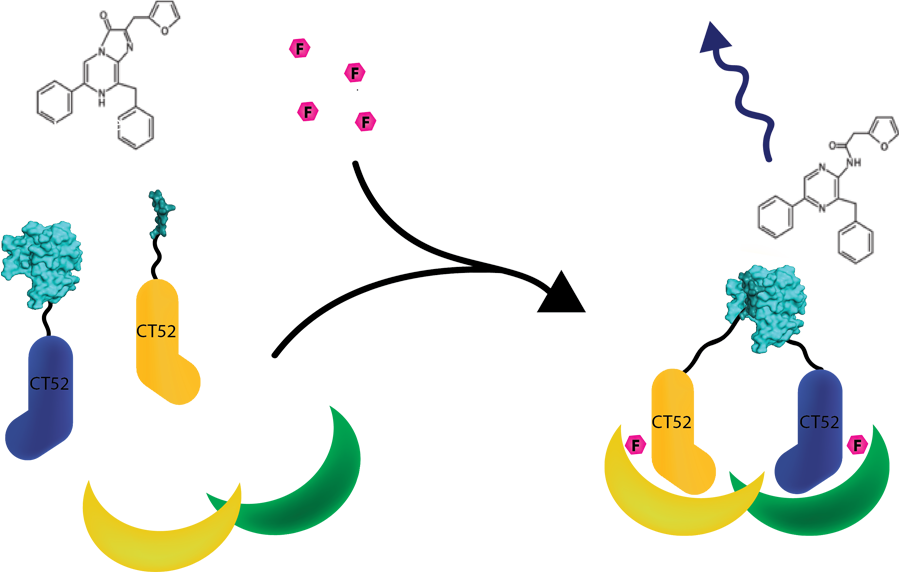
Figure 1: Measurement system for T14-3-3 heterodimers using the NanoBiT system linked to CT52. Under the influence of fusicoccin, the split NanoBits attached to CT52 will bind to T14-3-3 and emit a blue fluorescence light when dimerized and when furimazine is present.
Bioluminescence confirmation
To determine whether the split luciferase system is working, a bioluminescence signal should be visible around 460nm when SmallBiT and LargeBiT dimerize. When Furimazine is added to the dimerized Nanoluc luciferase, the Furimazine is converted into Furimamide. When this reaction occurs, light with a wavelength of 460 nm will be emitted. The spectrum of the CT52-NanoBiT system is visualed below.
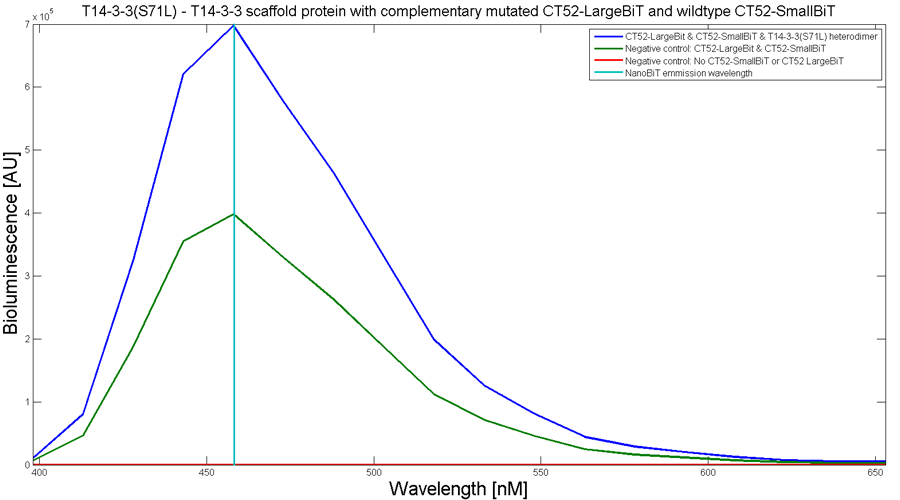
Figure 2: Bioluminescence spectrum with CT52(I947H)-LargeBiT and CT52-SmallBiT, assembled on T14-3-3(S71L) - T14-3-3
An example of a measurement which can be done on a heterodimer is a T14-3-3 heterodimer concentration gradient assay, in which the functionality can be verified. This measurement is shown below for T14-3-3(S71L) - T14-3-3. More measurements on T14-3-3 heterodimer scaffold with the CT52-NanoBiT system can be found on our wiki "http://2016.igem.org/Team:TU-Eindhoven/Results".
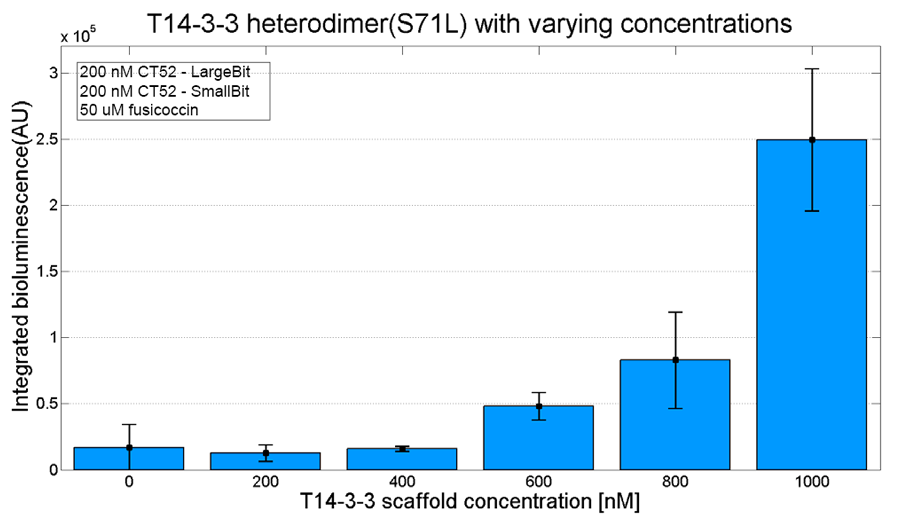
Figure 3: T14-3-3 functionality measurement with varying scaffold concentrations from 0 nM to 1 µM T14-3-3(S71L/I72V)-T14-3-3. 50 µM fusicoccin was used for all measurements. Mutant forms CT52 (I947F)-LargeBiT and CT52(wildtype)-SmallBiT were used with a concentration of 200 nM. The substrate used was 1250x diluted from stock.
NanoBiT calibration
In order to analyse the performance of our CT52 fused NanoBiT fragments we measured the luminescence they produce at varying concentrations from 10nM to 10mM.
After global optimization using metropolis Monte Carlo and Latin hypercube sampling the best fit resulted in LUmax = 3.5* 105 ±1.5 a.u. and KD = 996±163 μM (see figure 4), standard deviations are calculated with the Jacobian.
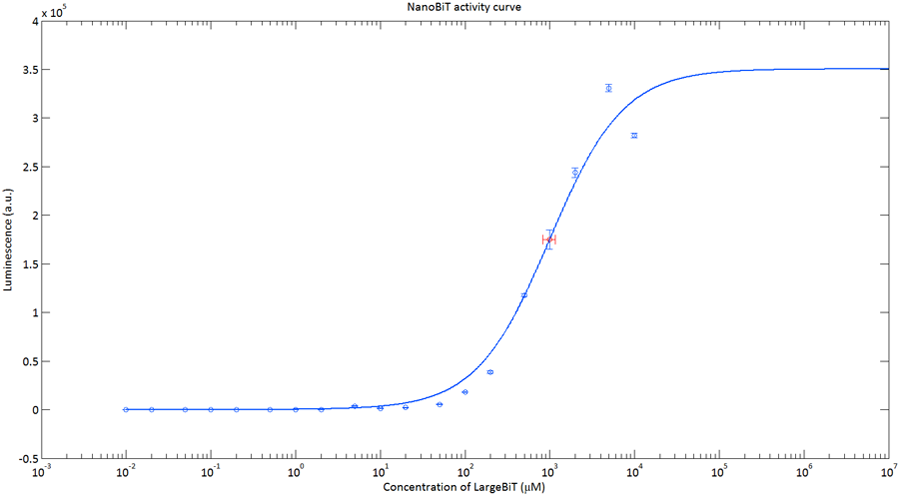
Figure 4:The Luminescence of NanoBiT as a function of the concentration of LargeBiT. The concentration of smallBiT is kept constant at 100 nM. The circles represent the measured datapoints, the errorbars represent the standard deviation of the measures (n = 2). The red circle and errorbars represent the dissociation constant and its standard deviation respectively.
Informational Contribution iGEM TU-Eindhoven 2017
The sequence has been determined and published by TU-Eindhoven 2017[http://2017.igem.org/Team:TU-Eindhoven]. The sequence does not include the ATG start codon. The end of the part contains a Stop codon, which makes this part suitable to place behind other BioBricks. The information of the sequence can allow people to acquire more insights on the functionality of the part.
References
[1] Kyle Hooper, "Applications of a smaller, brighter, more versatile luciferase: NanoLuc™ Luciferase Technology", Presentation slides Fall 2012. [3]
//function/reporter/light
| emission | 460 nm |