Part:BBa_K1321339
CBDcenA+Linker, RFC 25 standard
Cellulose binding domain (CBD) of Endoglucanase A (cenA) from Cellulomonas fimi with an endogenous C-terminal linker. The part is in Freiburg format (RFC 25) for ease of use in protein fusions.
Usage and Biology
The cellulose binding domain region occurs at the N-terminus of the cenA gene ([http://www.uniprot.org/uniprot/P07984 UniProt P07984]) and is from the [http://www.cazy.org/CBM2.html CBM family 2].
The linker sequence (PTTSPTPTPTPTTPTPTPTPTPTPTPTVTP) is Pro-Thr box which has an extended conformation and acts as a hinge region (1). The endoglucanase CBDcenA has 50% homology to the exoglucanase CBDcex (BBa_K863101) and the linker is highly conserved in both, but the order of the catalytic, linker and cellulose-binding regions is reversed (2). It has been shown that CBDcenA has the highest binding affinity for crystalline cellulose out of the C. fimi CBDs (3).
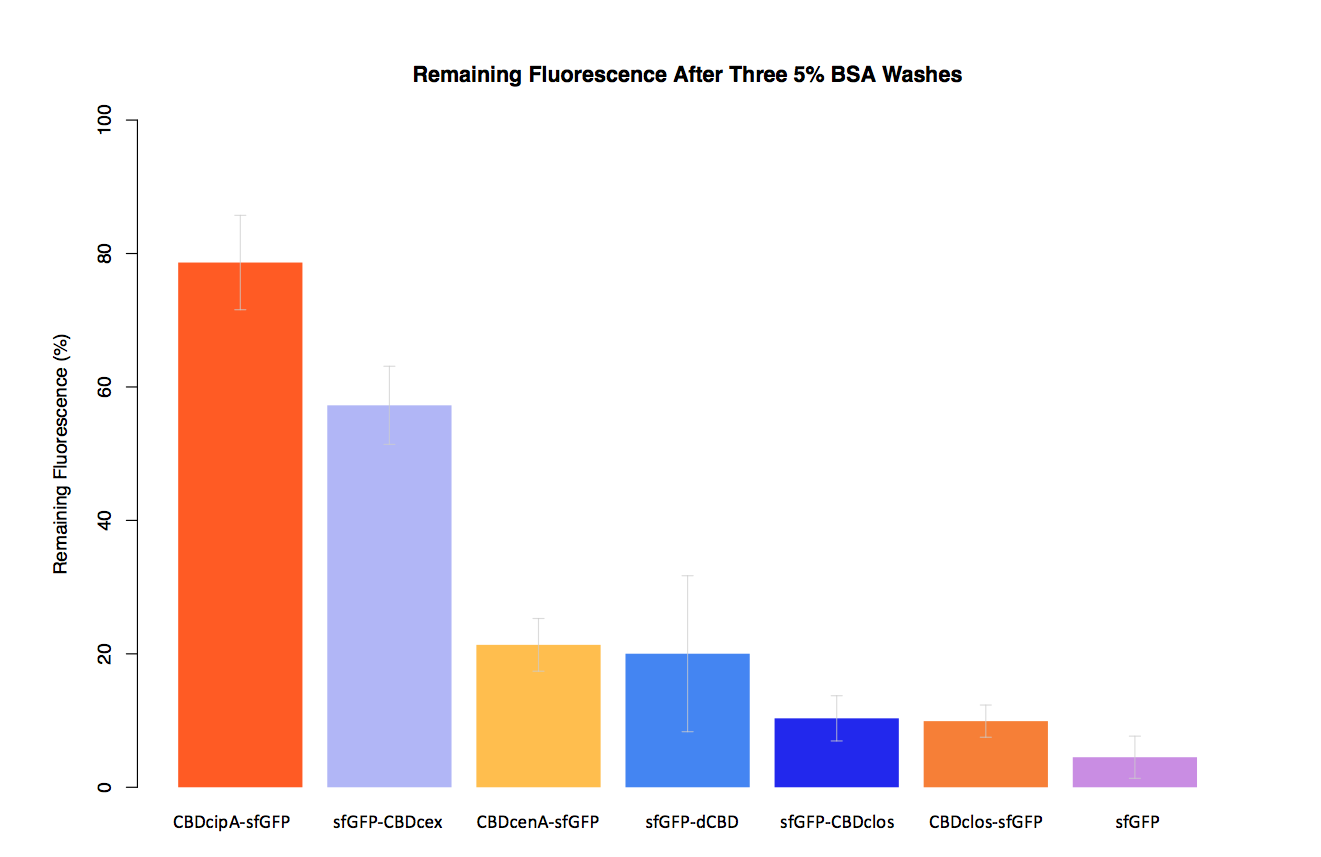
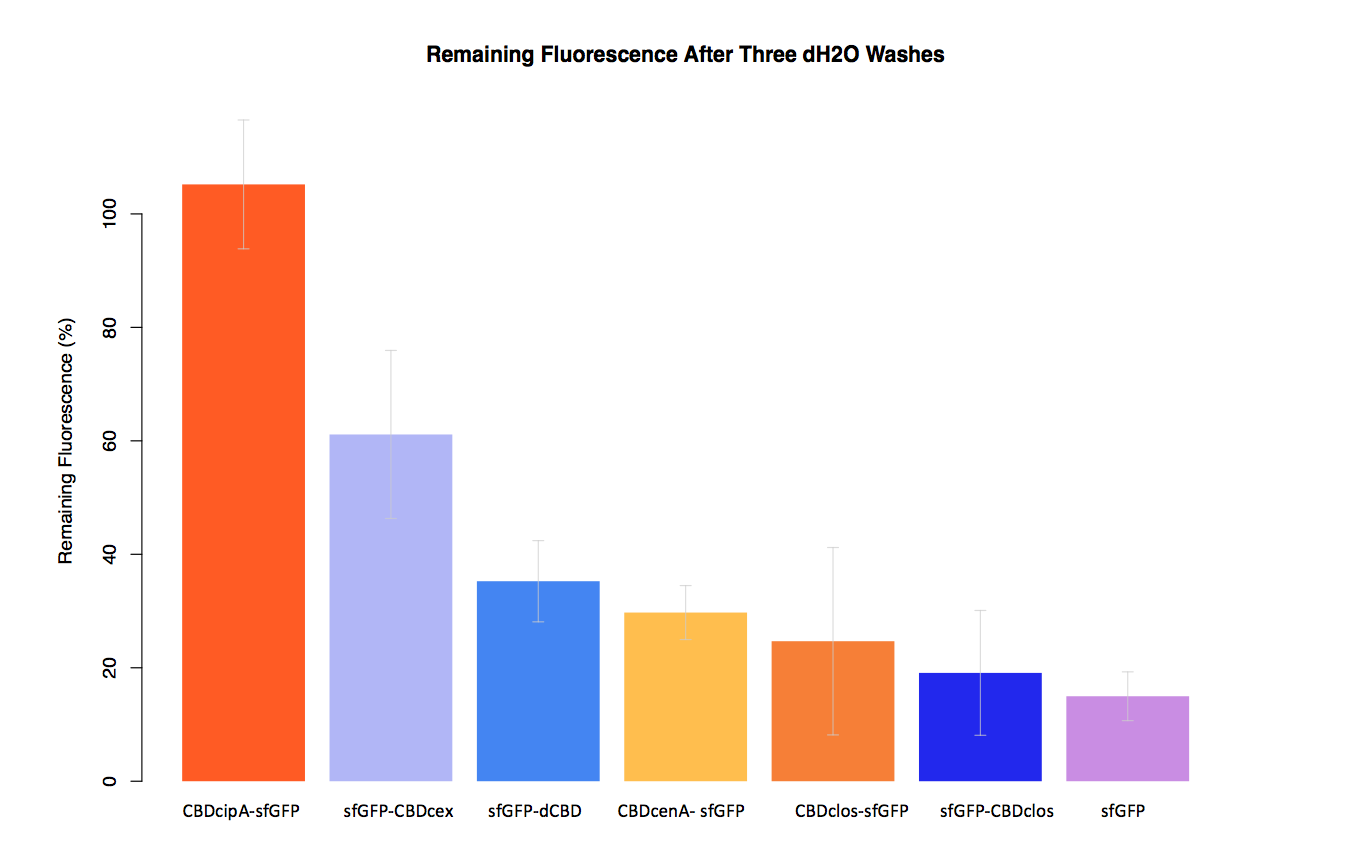
As part of our project we carried out an assay to determine the relative binding ability of CBD-sfGFP fusions to bacterial cellulose.
In our second assay performed to determine the relative strengths of various CBDs’ binding to bacterial cellulose – represented by the percentage fluorescence left from CBDs fused to (sfGFP (RFC25)) bound to bacterial cellulose discs, when subjected to various washes (protocol [http://2014.igem.org/Team:Imperial/Protocols here]) - it was determined that the fusion with CBDcenA had moderate binding ability in comparison to five other CBDs fused to sfGFP, having the third and fourth highest fluorescence remaining after three washes with 5% BSA and dH2O respectively.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| Protein data table for BioBrick BBa_K1321339 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25: (underlined part encodes the protein) ATGGCCGGC ... CCCACCGGTTAA ORF from nucleotide position 1 to 423 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
1. Gilkes, N.R., Kilburn, D.G., Miller, R.C. & Warren, R.A. (1989) Structural and functional analysis of a bacterial cellulase by proteolysis. The Journal of biological chemistry. 264 (30), 17802–17808.
2. Warren, R.A., Beck, C.F., Gilkes, N.R., Kilburn, D.G., et al. (1986) Sequence conservation and region shuffling in an endoglucanase and an exoglucanase from Cellulomonas fimi. Proteins. 1 (4), 335–341.
3. Kim, H.-D., Choi, S.-L., Kim, H., Sohn, J.H., et al. (2013) Enzyme-linked assay of cellulose-binding domain functions from Cellulomonas fimi on multi-well microtiter plate. Biotechnology and Bioprocess Engineering. 18 (3), 575–580.
| None |