Part:BBa_K1321090
Phytochelatin (PC) EC20 + Linker-dCBD
Phytochelatin metal binding peptide fused N-terminally to dCBD, which contains an endogenous N-terminal linker sequence.
This construct is part of a library of fusions with cellulose binding domains which we designed to bind to cellulose and enable capture of heavy metals ([http://2014.igem.org/Team:Imperial/Functionalisation project page]). Other fusion parts with this metal binding protein can be seen in the table below: 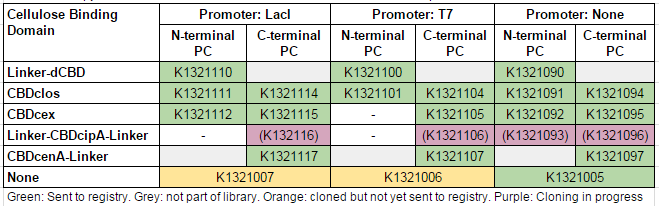
Note that the start and stop codon, plus 6 bp either side of the sequence, are included the RFC25 prefix and suffix which is not shown.
For reference the cellulose binding domain binding capability to bacterial cellulose was measured relative to other cellulose binding domains when fused to sfGFP, the data for which can be seen here (here) - K1321348.
As part of our project, we needed to assay the metal binding capability of our phytochelatins fused to CBDs (cellulose binding domains). The parts used for this assay are
Phytochelatin+CBDcex, Phytochelatin+dCBD, CBDcipA+Phytochelatin, Phytochelatin alone and sfGFP+dCBD wash (only one wash).
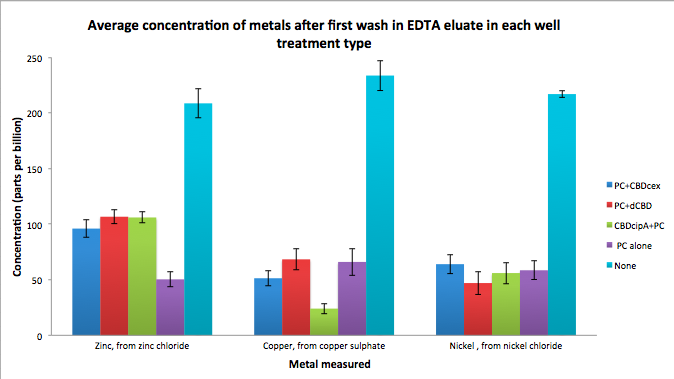
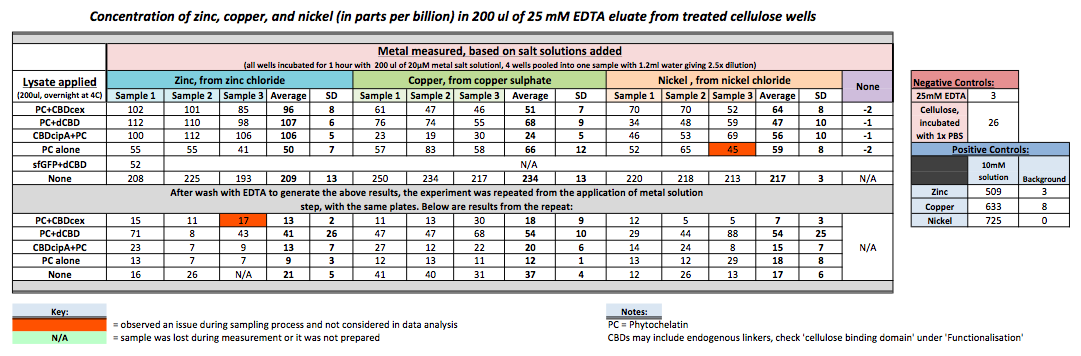
Phytochelatin fused to our CBDs were bound onto cellulose that were dried in the bottom of 96-well plates and tested against 3 different metals (nickel, copper, zinc). First, the fusion protein cell lysate was incubated overnight in the cellulose wells. Following this, the metal salt solutions are added in excess into the wells. Finally, an EDTA step removes the bound metal ions into solution, and the metal concentration in solution is quantified by mass spectrometer. Multiple washes with PBS and water were done between each binding step, ensuring that the metal ions that are measured were released from the phytochelatin.
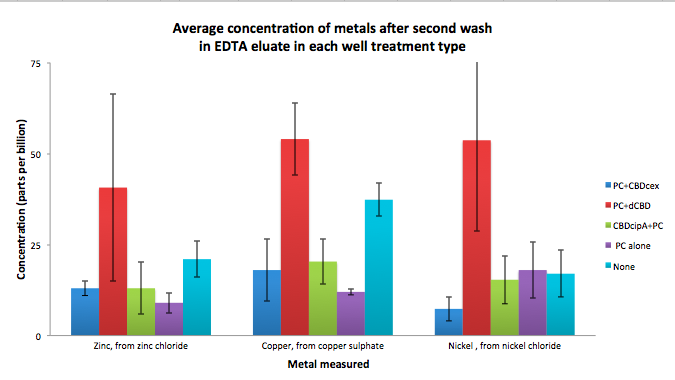
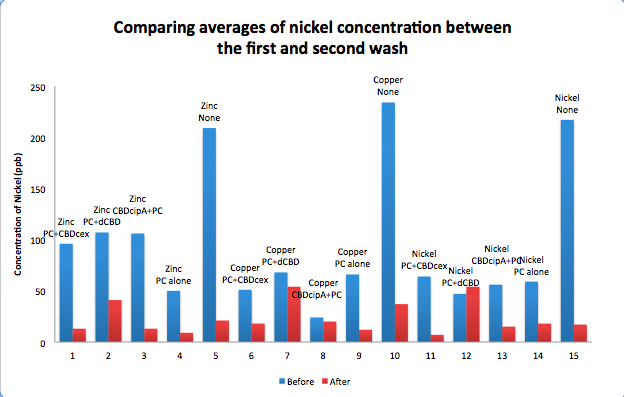
To evaluate if these CBD fusions were reusable, we re-applied metal ion solutions onto the same wells with the CBD fusions adhered. We washed the wells as before, then eluted with EDTA in the same manner. The results from the each elution is shown below, with the final graph comparing between the first and second wash. We found along the way our bacterial cellulose alone had some metal chelating properties, as was also confirmed by our filtering set up with the dCBD-phytochelatin, but the second elution seems to show less background noise as the EDTA disrupts cellulose's natural binding to the metal ions. The full table of results are shown below as well. The full assay protocol can be found as ‘Metal binding assay protocol’ at this link: http://2014.igem.org/Team:Imperial/Protocols
To read more about this assay and explanation of these results, please see:
http://2014.igem.org/Team:Imperial/Water_Filtration
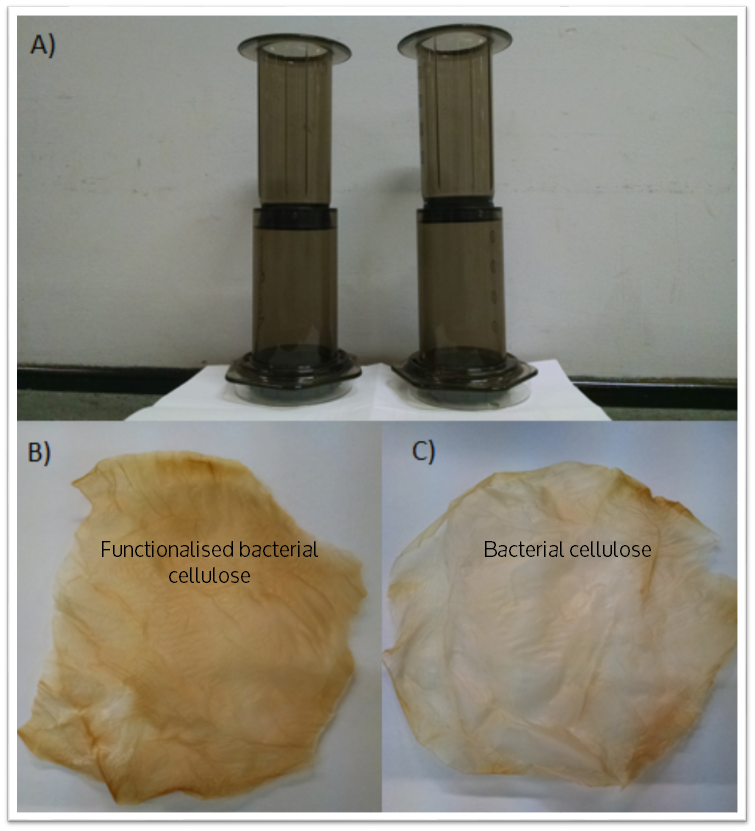
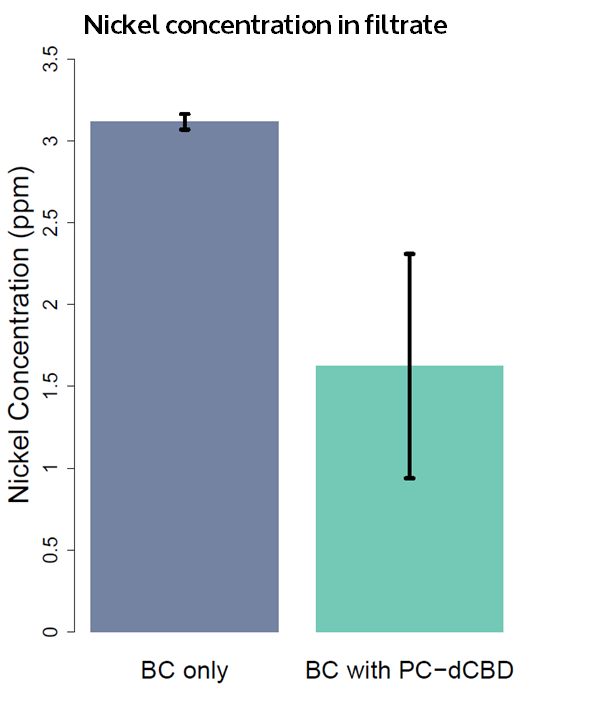
As a measure of the metal binding ability of Phytochelatin-dCBD attached onto bacterial cellulose, a filtration setup was developed and tested as displayed in the figure below. The setup includes a coffee press with a bacterial cellulose filter contained at the bottom supported by a 0.5 mm porous structure. A 50 mm diameter bacterial cellulose pellicle in the shape of a circle was used as the filtration membrane. Both pellicles were produced by ATCC 53582.
Cell lysate was prepared by inoculating a single colony of Phytochelatin-dCBD in 10 ml of standard LB media overnight under standard conditions of 37 C 180 rpm shaking. This pre-culture was used for a 1000 ml grow up of LB media, which was incubated overnight under the same conditions. This media was spun down in a centrifuge pot, sonicated and resuspended in 5 ml of PBS solution. The cell lysate was applied on to pasteurised bacterial cellulose with a spreader and dried at room temperature. For the control, a pellicle without any cell lysate applied was used.
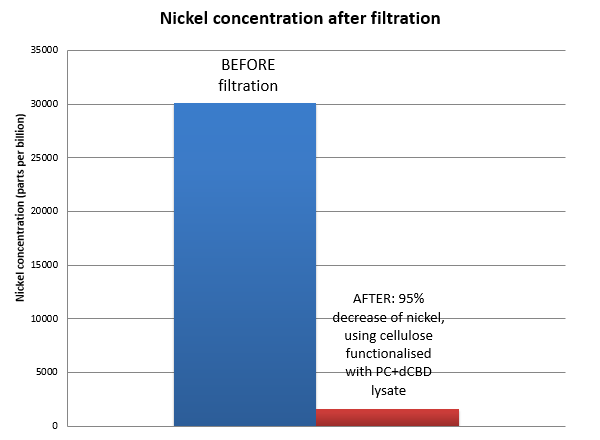
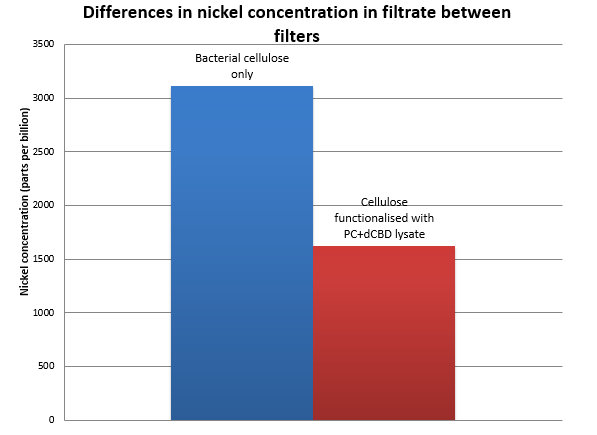
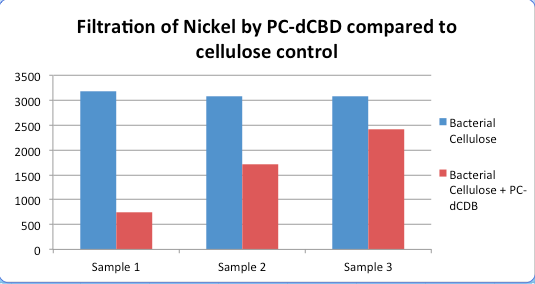
Results were obtained as shown in the figures below:
The filtering can be summarised as shown in the figure below, showing better Nickel binding abilities of the functionalised cellulose in comparison to the control.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |