Part:BBa_K1321348
sfGFP fused to dCBD with linker in RFC 25
Super-folder GFP fused N-terminally to dCBD (a cellulose-binding domain), which contains an endogenous N-terminal linker sequence.
This construct is part of a library of Super-folder GFP fusions with cellulose binding domains, which we used to assay the CBD binding affinity. Please see our [http://2014.igem.org/Team:Imperial/Functionalisation project page] for more information. The collection of sfGFP-CBD fusion parts can be seen in the table below: 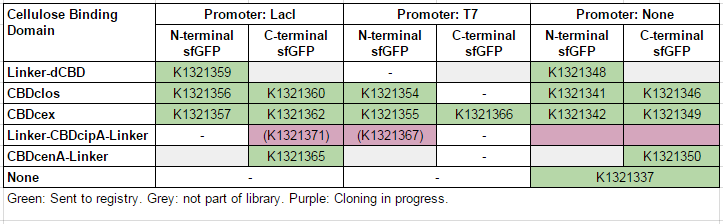
Note that the start and stop codon, plus 6 bp either side of the sequence, are included in the RFC25 prefix and suffix which is not shown.
As a part of our project, we carried out an assay to determine the relative binding ability of CBD-sfGFP fusions to bacterial cellulose.
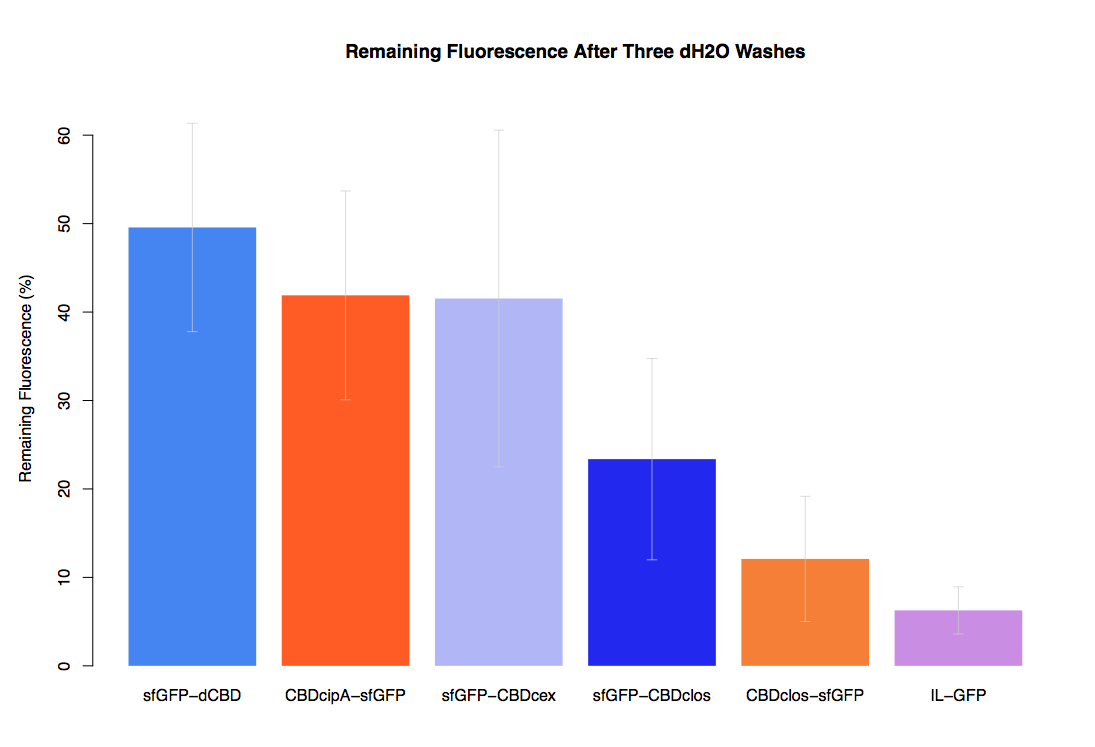
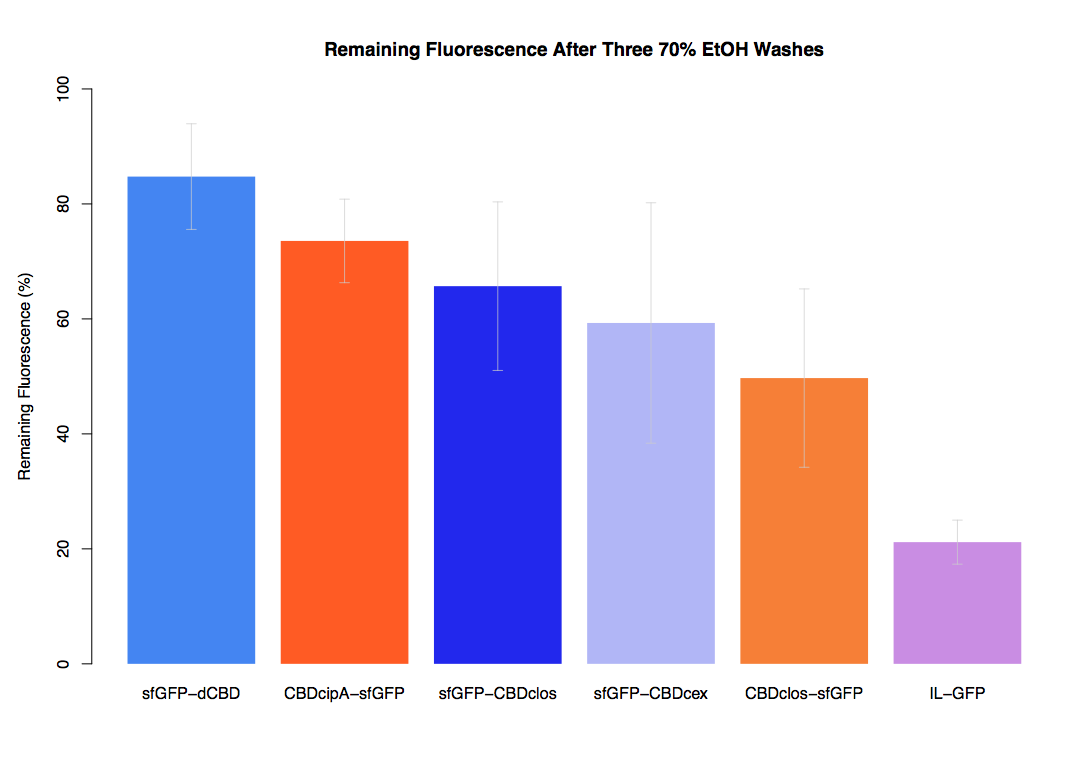
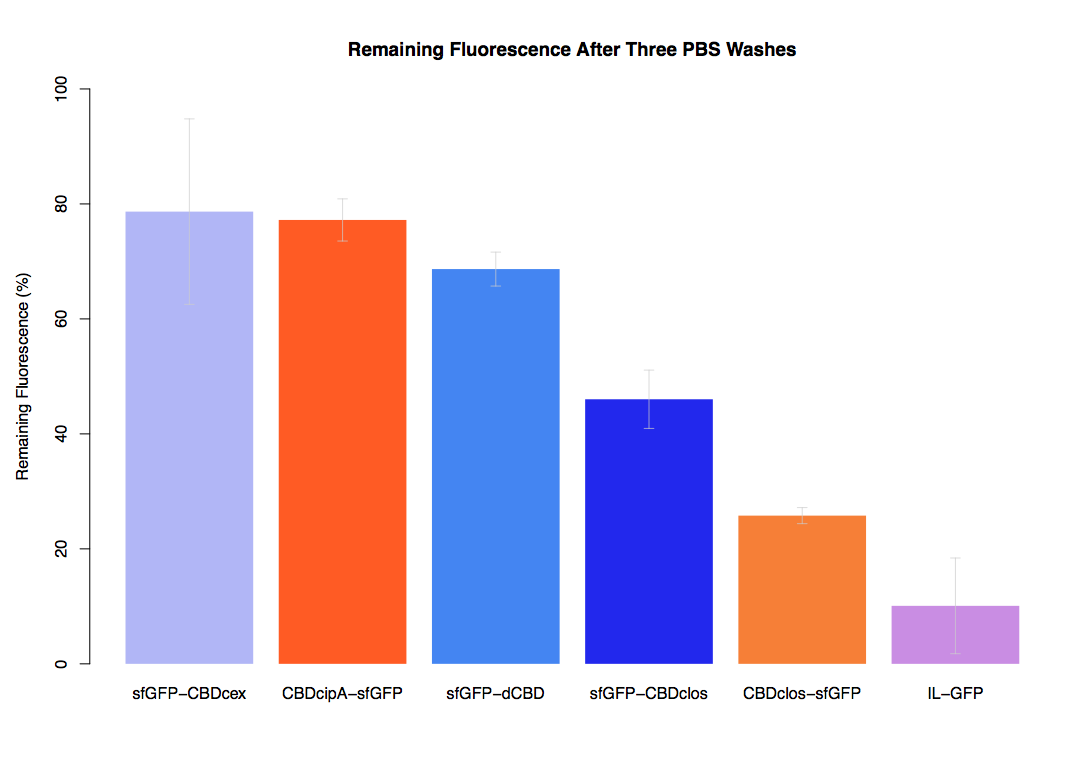
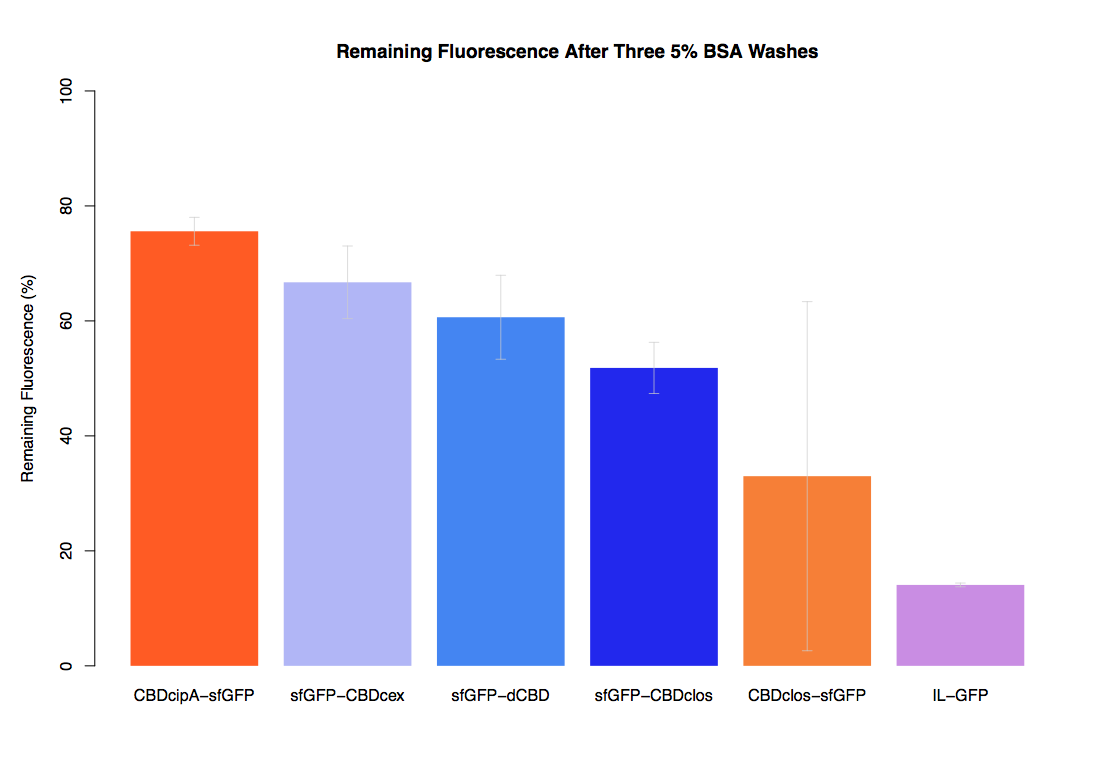
In our first assay performed to determine the relative strengths of various CBDs’ binding to bacterial cellulose – represented by the percentage of fluorescence left from CBDs fused to (sfGFP (RFC25)) bound to bacterial cellulose discs, when subjected to various washes - it was determined that the fusion with dCBD had the greatest binding ability in comparison to four other CBDs fused to sfGFP after three washes with both dH2O and 70% EtOH.
In the same assay, results suggested that dCBD had on average the third greatest ability to bind bacterial cellulose, after three washes with both PBS and 5% BSA.
Characterization: BrownStanfordPrinct 2019
Group: BrownStanfordPrinct 2019
Authors: Cale Lester
Summary: We measured the fluorescent activity of BBa_K1321348 while bound to bacterial cellulose after lyophilization.
Documentation:
We uniformly cut 1cm by 1cm squares of bacterial cellulose paper which were soaked in either control uninduced cell lysate, control deionized water, or induced cell lysate containing BBa_K1321348 for 24 hours while shaking at room temperature. The squares were then lyophilized for 24 hours and subsequently inserted flat into the bottom of wells in a 96 well plate where fluorescence was measured using sGFP's optimal excitation of 485nm and emission of 510nm.
As shown in the graph, the functionality of the fusion protein is retained even after lyophilization, and is significantly greater than the fluorescence shown in either of the negative controls even after lyophilization. Qualitatively, the cellulose binding activity is shown to be retained. The error bars represent the standard error measured as the standard deviation of the mean divided by the root of the sample size. None of the error bars overlap, so we can be confident that these results are statistically significant (n=6).
Contribution: WHU-China 2019
Group: WHU-China
Authors: Junfan chen
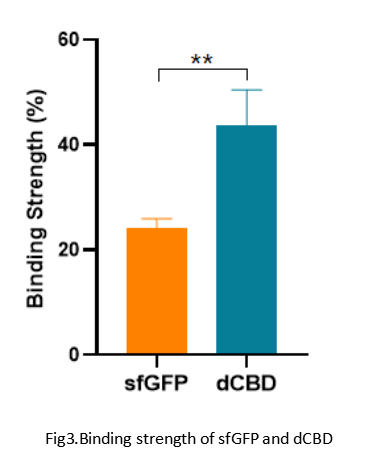
Summary: We confirmed the function of dCBD fused with sfGFP as a reporter by testing the combination strength between dCBD and bacteria cellulose.
Documentation:
1-Qualitative experiment
The experimental group we set is the mixture of dCBD sample with bacteria cellulose homogenate. We also set a negative control which mixed sfGFP with bacteria cellulose homogenate.
After a series of operations such as mixing, centrifugation and washing, the following conclusions were obtained:
When we used ddH2O (0.1% TWEEN 20) to wash the mixture and centrifugated it, we got that the residual amount of sfGFP on BC gel was lower than that of dCBD which was visible to naked eyes.
1-Quantitative experiment
We did the quantitative test of fluorescence intensity by using a multifunctional microplate reader. We got the results as follows:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//cds/reporter/gfp
| None |