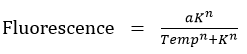Part:BBa_K1184000
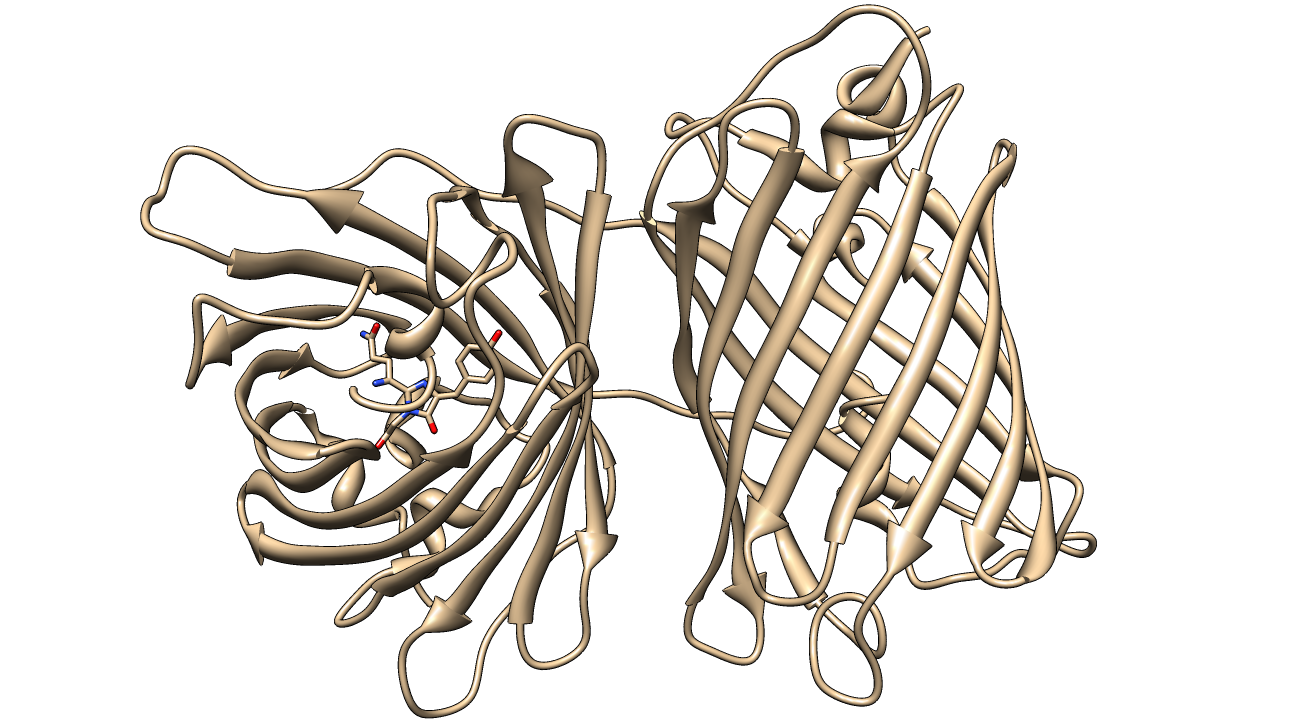
KillerRed
KillerRed is a red fluorescent protein that produces reactive oxygen species (ROS) in the presence of yellow-orange light (540-585 nm). KillerRed is engineered from anm2CP to be phototoxic. It has been shown that KillerRed produces superoxide radical anions by reacting with water. Superoxide reacts with the chromophore of KillerRed, causing it to become dark, which ultimately gives rise to a bleaching effect. KillerRed is spectrally similar to mRFP1 with a similar brightness. KillerRed is oligomeric and may form large aggregates in cells. Expression of KillerRed and irradiation with light may act a kill-switch for biosafety applications.
Superoxide (O2•-) is the radical anion of molecular oxygen, which can react with protein side chains and lipids to harm cells.
This sequence is codon optimized for mammalian cells and has 13 rare proline codons for E. coli (CCC) and one rare arginine codon (AGA). Codon optimization should be taken into consideration if large amounts of the protein are required. Expression on a high-copy plasmid has produced detectable fluorescent signal, however.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 151
Illegal BsaI.rc site found at 442
Characterization
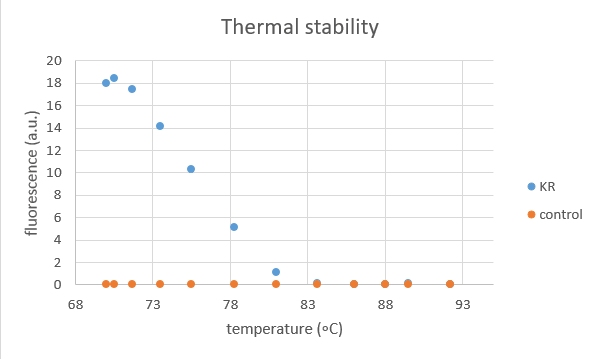
2016 NYMU-Taipei team characterized this part with thermal stability assay.
KillerRed protein was extracted from transformed E.coli and a control group of empty vector was set to eliminate the influence of other proteins.
This test aims to show the temperature influence on KillerRed protein by measuring its fluorescence after being treated for 2 hours at the following temperatures: 70, 70.5, 71.7, 73.5, 75.5, 78.3, 81, 83.6, 86, 88, 89.5, 92.2 (degree Celsius).
- It is obvious that KR contribute to most of the fluorescence.
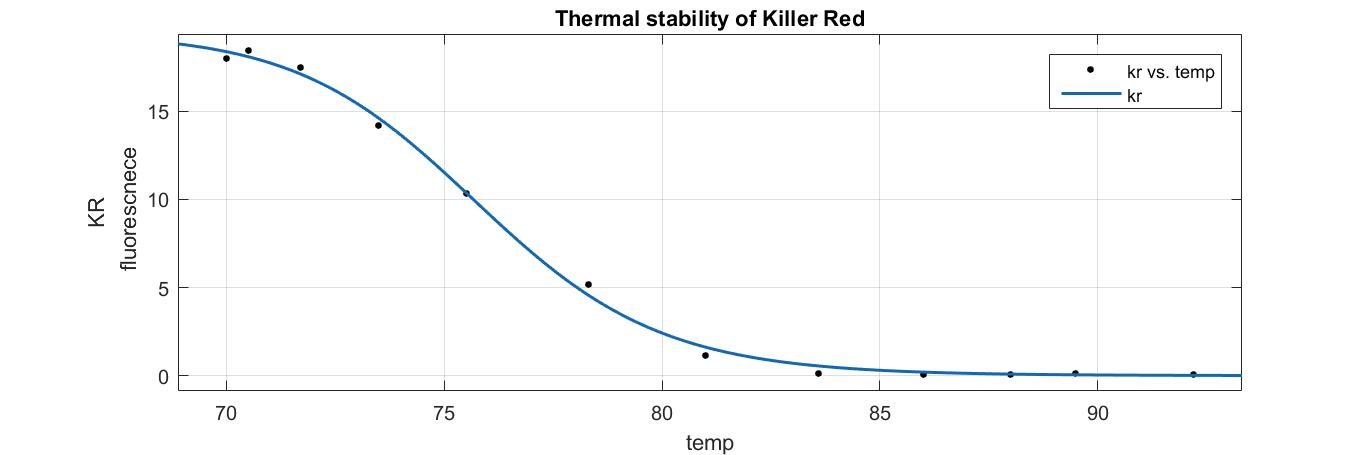
- A sigmoidal curve was fitted to find the particular temperature K at which half of the KR protein is denatured studied by measuring the fluorescence.
a=19.42
n=36.1
K=75.79
R-square=0.998
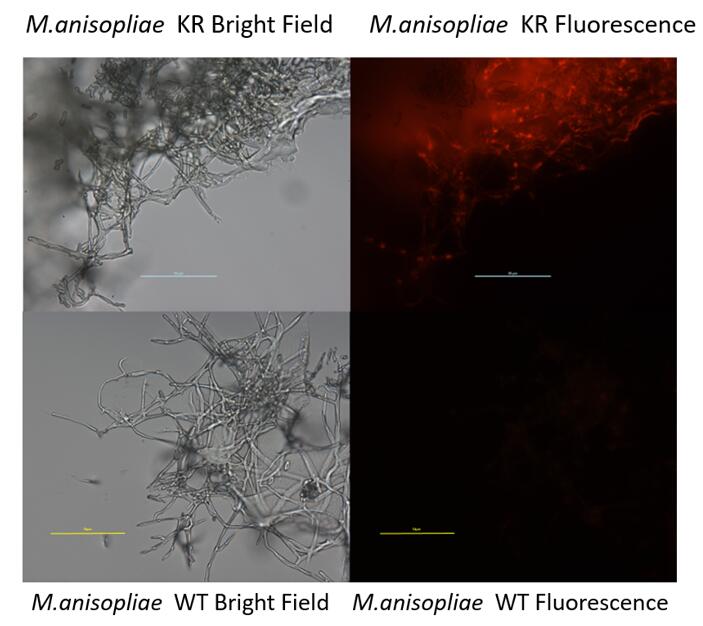
NYMU 2016 also developed this part to construct a fungal expression cassette. (BBa_K2040121)
- This new device was successfully expressed in M.anisopliae
2019 KOREA team characterized this part with phtotoxicity
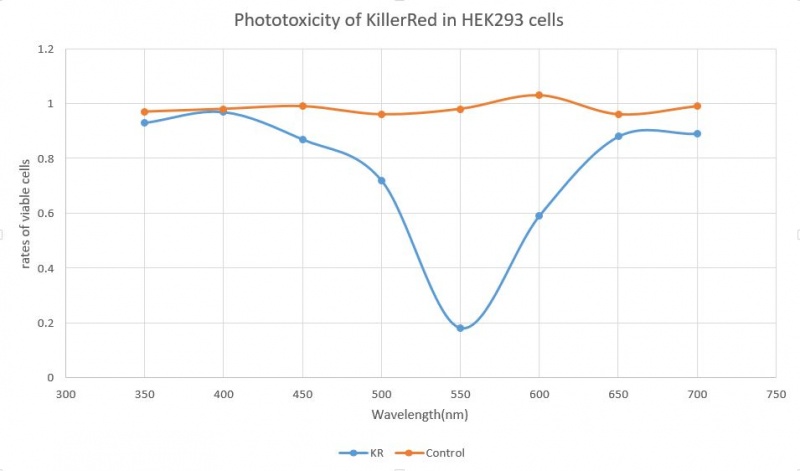
KillerRed is the light-induced killswitch producing rhe reactive oxygen spieces. When cells transfected with killswitch gene is exposed to the green or orange light(540-590nm), the KillRed becomes toxic and kills the cell. The normalized excitation of fluorescence has a peak at near 600nm wavelength.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
2023 Hangzhou-SDG Team characterized this part as a part of of a suicide switch
KillerRed can be a part of the suicide switch for M. anisopliae when being connected after the hemolymph inducible promoter Pmcl1/Pmcl1(short).
Larvae of Galleria mellonella were killed by engineered M. anisopliae. The corpses were placed under sunshine for days until spores formed outside the insect body. Spores were collected by vertexing and the concentrations of spore suspensions were checked under a microscope.

Results showed that suicide switches consisting of KillerRed led to significantly fewer spores than WT did.
2023 ZJU-China Team characterized this part as a kill switch in Agrobacterium tumefaciens
KillerRed serves a crucial role in preventing Agrobacterium tumefaciens from inadvertently escaping into the environment.
In our experiments, we coupled the KillerRed protein with promoter 50Spro(BBa_K4628022) and terminator BBa_B0015. These elements were cloned into plasmid pGreenII and subsequently transformed into Agrobacterium tumefaciens GV3101 for further examination.
Result:
our plate light experiment demonstrated a noticeable reduction of engineered bacteria under natural light irradiation, regardless of whether the conditions were sunny or cloudy.

FIG. Results of light experiments of Agrobacterium tumefaciens with KillerRed V.S. empty plasmid
Considering that plants predominantly absorb blue-violet and red light, while reflecting light near 540nm (green). KillerRed emerges as an ideal biosafety ‘kill switch’. This is particularly effective when the light exposure is either sufficiently intense or prolonged.
2024 HZAU-China Team characterized this part as a a suicide switch in Bacillus subtilis
In the experiments, we coupled the KillerRed protein with the Pxyl promoter (BBa_K143014) and SpovG RBS (BBa_K143021). These elements were cloned into plasmid pHT315 and subsequently transformed into Bacillus subtilis 168 for further examination.

Result:
Our experiments with bacterial solutions under different colors of light show that KillerRed has different excitation sensitivity to different colors of light and is most sensitive to green light.

Our experiments on bacterial solutions under artificial green light and natural light show that KillerRed has a very obvious killing activity in Bacillus subtilis, and this effect is mainly caused by green light excitation.

//biosafety/kill_switch
//cds/reporter/rfp
//chassis/prokaryote/ecoli
//function/celldeath
//function/reporter/fluorescence
| color | Red |
| control | BBa_E1010 as a negative control |
| emission | 611 nm |
| excitation | 545-585 nm |
| function | Phototoxicity |
| origin | Engineered from anm2CP |
| output | Superoxide radical anion |