Part:BBa_K2201262
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) with an amber stop codon at amino acid po
The Ribulose 1,5-bisphosphate carboxylase oxygenase (RuBisCo) converts ribulose 1,5 bisphosphate to 3-phosphoglycerate. For the translational incorporation of a fluorescent noncanonical amino acid the codon for the amino acid at position 111 of the small subunit has been replaced with the amber stop codon TAG.
Usage and Biology
Ribulose 1,5-bisphosphate carboxylase oxygenase (RuBisCo) catalyzes the incorporation of inorganic CO2 to ribulose 1,5‑bisphosphate to form two 3‑phosphoglycerate molecules. The catalyzed reaction is shown in Figure 1.(Andersson 2008).
Due to its numerous side reactions, for example the oxygenase activity resulting in the production of 2‑phosphoglycolate when O2 is present, RuBisCo is a very inefficient catalyst. CO2 and O2 are competitive substrates in the two reactions and only the production of 3‑phosphoglycerate leads to CO2 fixation. [Andersson 2008, Jordan 1981]. For a more complex description please refer to iGEM Bielefeld 2014 BBa_K1465202. Our aim was to localize the RuBisCo with the help of the fluorescent noncanonical amino acid L-7-Hydroxy(L-coumrayl)ethylglycine(CouAA). To incorporate this amino acid during translation we used the orthogonal tRNA/aaRS BBa_ K2201204 that incorporates CouAA in response to the amber stop codon TAG. We designed seven different variants of the RuBisCo with one to three amber stop codons at different positions to find out how many fluorescent amino acids are necessary to localize the RuBisCo and if the position of the fluorescence amino acid influences the fluorescence signal. The constructed variants are listed below. All these variants are also available as composite parts under control of a T7-promotor behind the carboxysome to colocalize the RuBisCo and the carboxysome, which is labeled with the green fluorescent protein (GFP). The numbers of the composite parts are listed behind the number of the basic part.
- BBa_K2201261 (BBa_K2201361) with a TAG codon at position 2 of the small subunit
- BBa_ K2201262 (BBa_K2201362) with a TAG codon at position 111 of the small subunit
- BBa_K2201263 (BBa_K2201363) with a TAG codon at position 474 of the large subunit
- BBa_K2201264 (BBa_K2201364) with a TAG codon at position 2 and psition 11 of the small subunit
- BBa_K2201265 (BBa_K2201365) with a TAG codon at position 2 of the small and position 474 of the large subunit
- BBa_K2201266 (BBa_K2201366) with a TAG codon at position 11 of the small and position 474 of the large subunit
- BBa_K2201267 (BBa_K2201367) with a TAG codon at position 2 and 111 of the small and position 474 of the large subunit
To compare the localization with the fluorescent amino acid to the localization with a fluorescent protein we constructed a fusion protein containing the RuBisCo and a red fluorescent protein (mRFP) BBa_K2201260 . Like the other variants it is also available as composite part unter control of a T7-promotor behind the carboxysome(BBa_K2201360).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1375
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 818
Illegal BamHI site found at 1499 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
To colocalize the RuBisCO in vivo, a plasmid containing the CDS a carboxysome-GFP fusion protein and one of the RuBisCO variants containing the amber stop codon was cotransformed with BBa_K2201204, containing the CouAA-aaRS and tRNA in pSB3T5. The seven different variants and the negative control were prepared like described before for the CLSM images. The images of the negative control and the variant containing one CouAA at amino acid position 2 of the small subunit are shown in Figure 2. The construct BBa_K2201360 containing the fusion protein of mRFP and RuBisCO showed no visible fluorescence. Maybe the linker was not suitable, so the mRFP was not able to fold correctly.

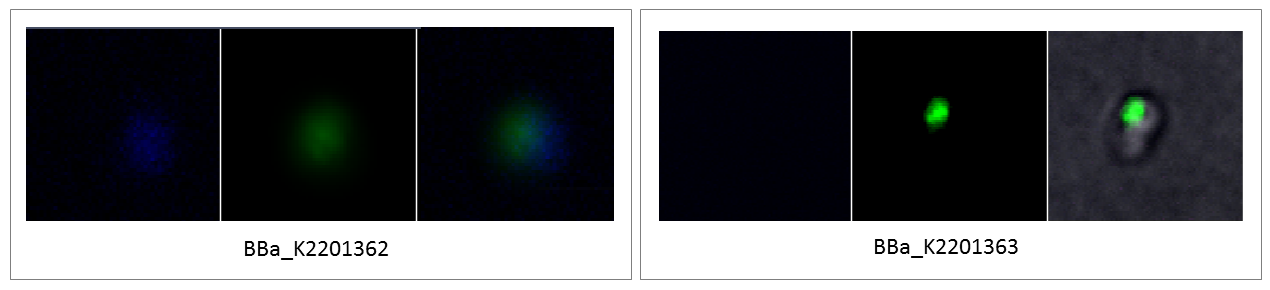
Unlike Sup35, the RuBisCO is not localized in the whole cytoplasm. It seems like most of the RuBisCo is localized in the carboxysome and only a small amount is localized over the whole cell. The fluorescence image of BBa_K2201362 (stop codon at position 111 of the small subunit) shows the same localization of the RuBisCO inside the carboxysome. Furthermore, there is no visible difference in the fluorescence intensity between BBa_K2201361 and BBa_K2201362 (Figure 2 and 3). In contrast, BBa_K2201363 (CouAA at position 474 of the large subunit) shows no visible fluorescence (Figure 3). This indicates that the position where CouAA is incorporated has a strong influence on the fluorescence signal, like assumed by our expert Prof. Nediljko Budisa.
Furthermore, we wanted to investigate if the incorporation of more than one fluorescent amino acid strengthens the fluorescence signal. Therefore, we prepared the four RuBisCO variants containing more than one amber stop codon like described before for fluorescence microscopy. The resulting images are shown in figure 4.
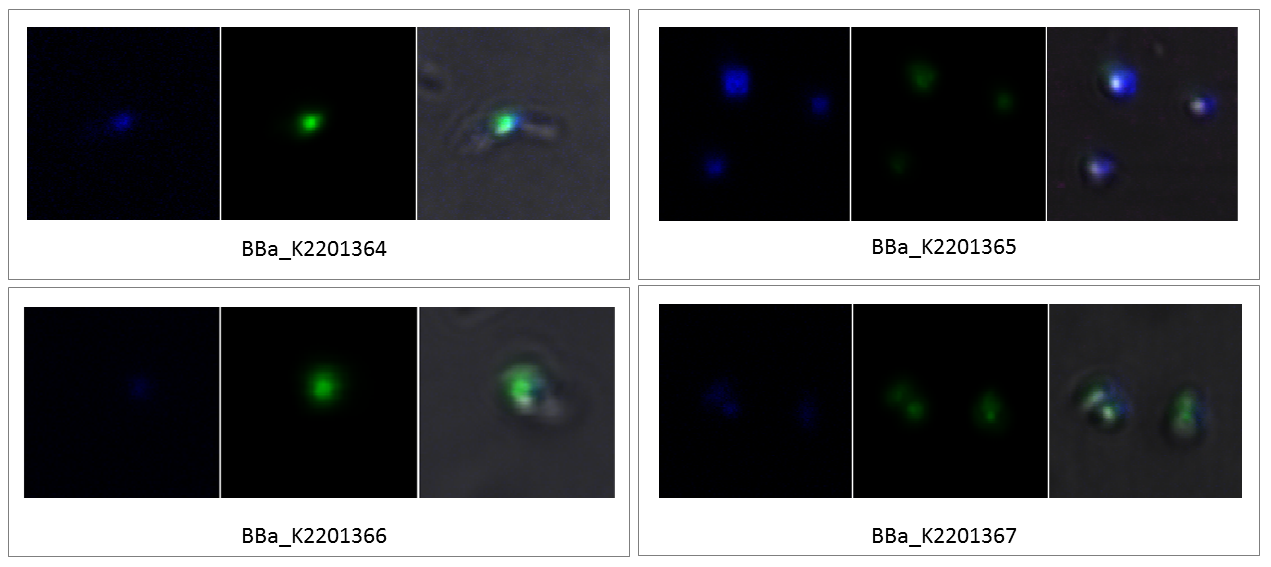
Compared to the fluorescence of the samples containing only one amber codon (BBa_K2201361, BBa_K2201362 and BBa_K2201363), and containing all three amber codons, (BBa_K2201367) the fluorescence of the samples containing two amber codons seemed to be the strongest. In all three of the samples containing two amber codons, and thus two CouAAs, the localization of the RuBisCO inside the carboxysome is visible. Another amber codon more could not increase the fluorescence intensity. This could be a result of a lower yield or the relatively low fluorescence of CouAA incorporated at position 474 of the large subunit.
All in all, the position of the CouAA and the number of the incorporated amino acids influence the fluorescence intensity and thus the quality of the localization in vivo. For the RuBisCo the incorporation of two CouAA resulted in the best fluorescence microscopy pictures. We proved that our tool is suitable for the localization in vivo and we were able to localize the RuBisCo inside the carboxysome.
| None |

