Part:BBa_K3385030
CRISPR_racA_KO
Theoretical expectation: racA encodes a Rho GTPase which controls actin dynamics and it is thereby also involved in polarity regulation. The knockout mutant was expected to have a hyperbranched morphology.
Full guide construct for knockout of the Rho GTPase gene (racA) involved in regulation of cell polarity in A. niger. This BioBlock has to be cloned into the PacI/Nt.BbvCI digested pFC330 backbone.
Functionality: The sgRNA efficiency has been accessed through the technique to assess protospacer efficiency (TAPE) [2]. A repair oligo is used to mediate homologous recombination, where a highly efficient sgRNA will show no colonies without the repair oligo, while less efficient sgRNA will show a reduced number of colonies.

Results: Below is a picture showing A. niger transformed with CRISPR_racA_KO and the repair oligo for racA. It shows efficient gene deletion when it's transformed with a repair oligo.

To see if the K/O’s were successful, other than looking at macromorphology, tissue PCRs were performed. By the amplification of specific primers, upstream and downstream of the gene, it can be verified if the gene has successfully been knocked out. If it has been knocked out the primers are gonna be closer to each other resulting in a smaller band in the Tissue PCR. However if the gene is still present in the genome, the band size will be the same as the target gene as seen in the table below.
| Targeted gene | Expected gene length after K/O | Control lenght |
|---|---|---|
| ΔchsC | 704 bp | 1867 bp |
| ΔaplD | 590 bp | 3807 bp |
| ΔracA | 709 bp | 1920 bp |

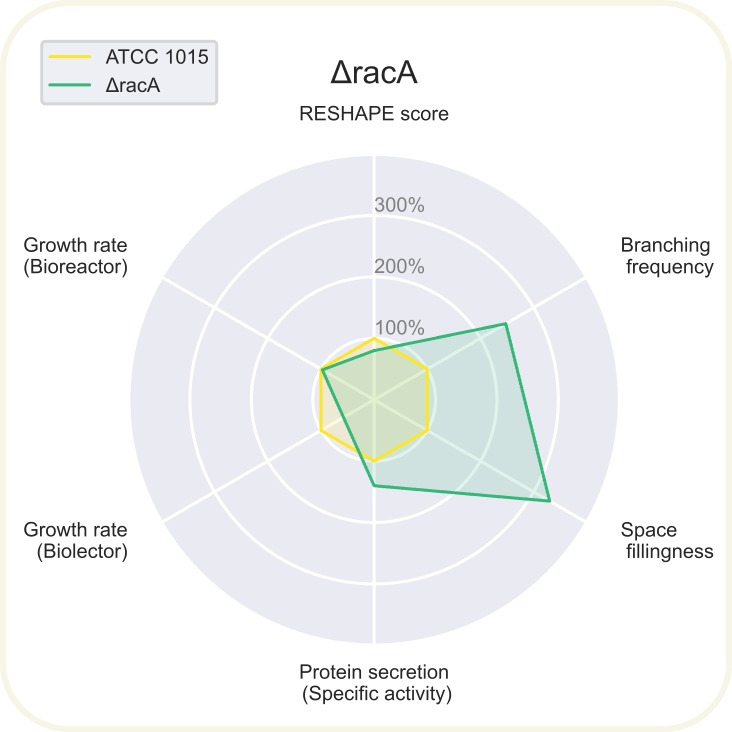
Radar chart showing 6 different parameters of ΔracA normalized to the reference values from ATCC 1015 (shown in yellow). Read about the axis in the summary section on the result page.
Plates
The strain was grown on Yeast Extract Peptone Dextrose (YPD), Transformation Media (TM), Creatine Sucrose Agar (CREA) and Czepek Yeast Extract Agar (CYA).
Microscope pictures and Simulation model

Left: Confocal microscope picture of ΔracA at 10X magnification after app. 24h growth. Right: Growth simulation for 12h performed using the Mycemulator.
Microscopic images were analyzed by the image analysis tool extracting growth parameters which were then fed to the Mycemulator. A simulation of ΔracA growing for 12 hours (using experimental growth rate from our BioLector data) is seen above.
Parameters specific for simulating ΔracA:
- Branching frequency: 0.0426064
- Gamma distribution parameters used for curvature angles: (0.8423059, 16.2365375)
- Beta distribution parameters used for branching angles: (1599535.5508791, 2.3452173)
- Experimental growth rate: 0.15865
BioLector
Comparing the growth kinetics of the ΔracA mutant with the reference strain ATCC 1015 in the BioLector, the mutant exhibits a longer lag phase and an almost double as long exponential growth phase. The growth rate for the mutant is much lower than for the reference strain, at μMax0.16h-1 versus μMax0.28h-1.
Growth profile of ΔracA over 72 hours, measuring absorbance at 620 nm. Plotted against the reference strain (ATCC 1015) shown in yellow.
Bioreactor
Looking at the growth of the ΔracA mutant in bioreactors, it showed an equivalently long lag phase as the reference strain ATCC 1015 followed by a slightly shorter exponential growth phase. ΔracA also has a growth rate almost equivalent to the growth rate of the reference strain, at μMax0.36h-1 and μMax0.32h-1 versus μMax0.37h-1 and μMax0.33h-1. Glucose levels start decreasing two thirds of the way through growth but are stil not completely depleted at the time of the last sample.
Left axis: Growth curve obtained from the off-gas analysis (CO2-44 (%)) of ΔracA plotted against the reference strain (ATCC 1015) in a logarithmic scale. Right axis: Glucose consumption by ΔracA during the fermentation in g/L (dark green).

Light microscope picture of ΔracA after 23 hours of fermentation with a 20X magnification.
The microscope picture from the bioreactor sample for ΔracA showed a pelleted morphology making it hard to compare with the brightfield microscopy pictures seen above. It showed a very hyperbranched pelleted morphology. When run in the bioreactor, the pelleted morphology was observable on a macroscopic level.
Protein secretion
The glucoamylase activity increases over time during the fermentation as seen in the figure below. The two duplicates showed a large difference in glucoamylase activity, one being equal to the reference strain whereas the other had an increased activity.
Glucoamylase activity in UA/mL of ΔracA from 8 samples taken during the fermentation. Plotted against the reference strain (ATCC 1015) shown in yellow.
A difference were also seen between the duplicates for the total protein secretion. Therefore, when calculating the specific activity both showed similar result with a value slightly higher than the reference strain. The difference the duplicates might be explained by a deviation in setting up the technical duplicates.

ΔracA bioreactor duplicates compared to reference strain (ATCC 1015) of the last time-point samples from the fermentations. Green: Glucoamylase activity in UA/mL. Blue: Specific activity in UA/mg calculated from the activity and the protein concentration. Purple: Protein concentration in mg/mL.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 489
Illegal EcoRI site found at 660
Illegal EcoRI site found at 831
Illegal SpeI site found at 978 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 489
Illegal EcoRI site found at 660
Illegal EcoRI site found at 831
Illegal NheI site found at 333
Illegal SpeI site found at 978 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 489
Illegal EcoRI site found at 660
Illegal EcoRI site found at 831 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 489
Illegal EcoRI site found at 660
Illegal EcoRI site found at 831
Illegal SpeI site found at 978 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 489
Illegal EcoRI site found at 660
Illegal EcoRI site found at 831
Illegal SpeI site found at 978
Illegal AgeI site found at 204 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 162
References:
[1] A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH. PLoS One. 2015 Jul 15;10(7):e0133085. doi: 10.1371/journal.pone.0133085. eCollection 2015. PONE-D-15-11561 [pii] PubMed 26177455
[2] Efficient Oligo nucleotide mediated CRISPR-Cas9 Gene Editing in Aspergilli. Nodvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH. Fungal Genet Biol. 2018 Jan 8. pii: S1087-1845(18)30004-5. doi: 10.1016/j.fgb.2018.01.004. 10.1016/j.fgb.2018.01.004 PubMed 29325827
