Part:BBa_K3191101
pBAD + AgrC + AgrA + GFP with Multiple Cloning Site
This part contains agrC and agrA genes from the agr system in Staphylococcus aureus. This system is responsible for detection of autoinducing peptides (AIP) using the AgrC protein. Interaction of AIP with AgrC causes AgrC to phosphorylate AgrA. Phosphorylated AgrA then initiates expression of genes downstream from the P2 promoter.
This part places agrC and agrA under the influence of a pBAD promoter. Downstream, a gene for GFP is found under the effect of a P2 promoter. This is designed so that AIP detection causes expression of GFP. A multiple cloning site was installed following the agrA gene for the insertion of additional BioBricks, such as SarA (BBa_K3191100).
University of Nebraska-Lincoln 2019
To characterize BBa_K3191101, cell cultures were grown alongside parts BBa_K1022100, BBa_K3191102, and BBa_K206000 cells. Cells were grown overnight and diluted to an OD of 0.05 in the morning with LB. The cultures were then allowed to grow to an OD 0D of 0.5. At this point, cells were washed with PBS and resuspended with 120 mM, pH 8.0 Tris-HCl. 30 mM EDTA and 10 mM of arabinose was then added to the cultures and the cells were incubated for 2 minutes. The cultures were then diluted with 2 mL of LB and antibiotics, and the cells were placed in a 37C shaker. Fluorescence was recorded every hour for three hours.
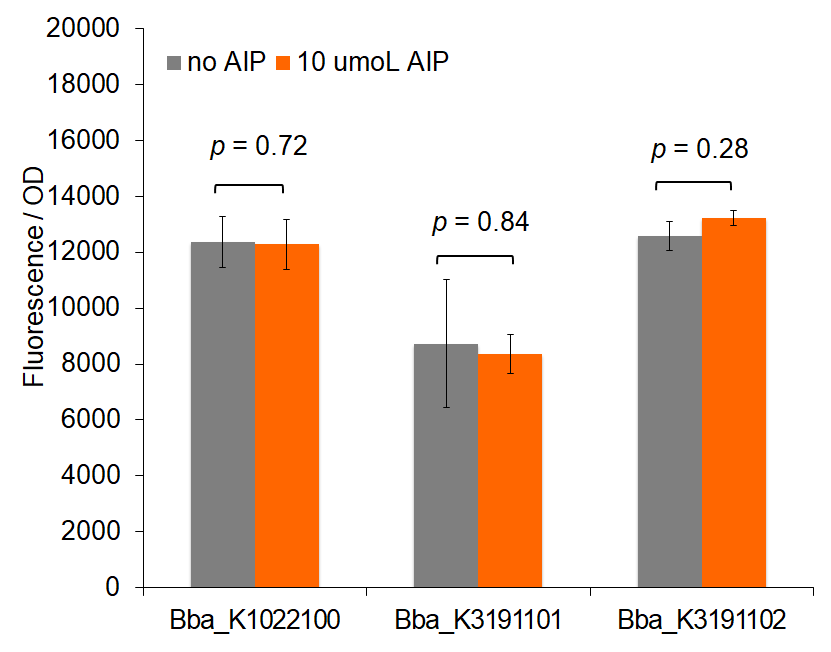
Figure 1: Total fluorescence of pre-permeabilized cells with and without AIP induction.
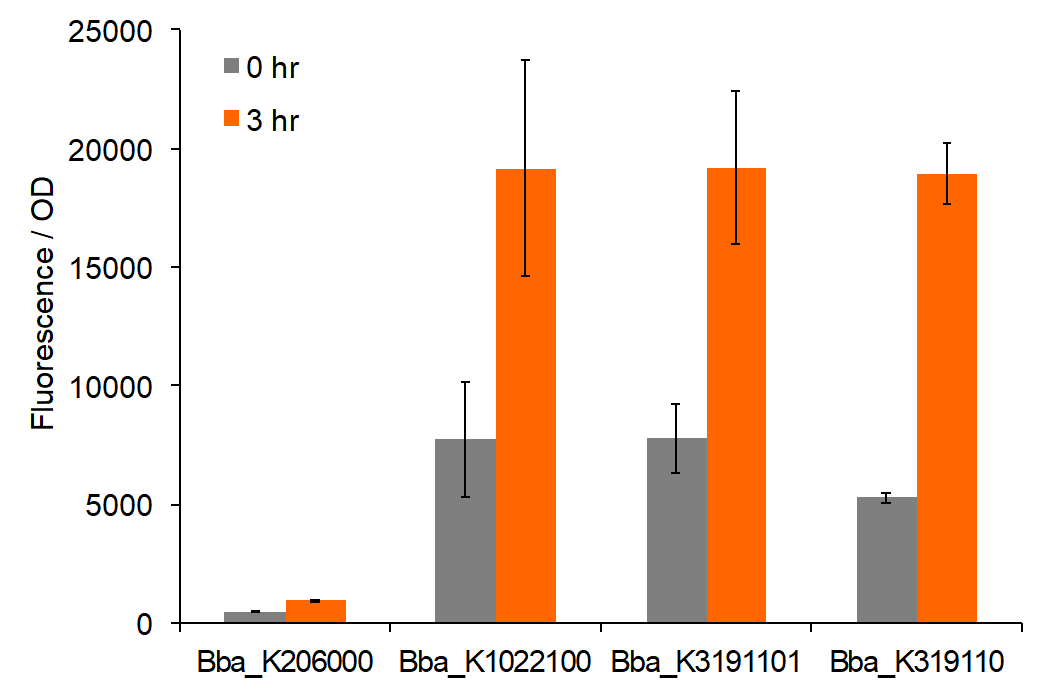
Figure 2: Total fluorescence of permeabilized cells at 0 and 3 hours post-induction with AIP.
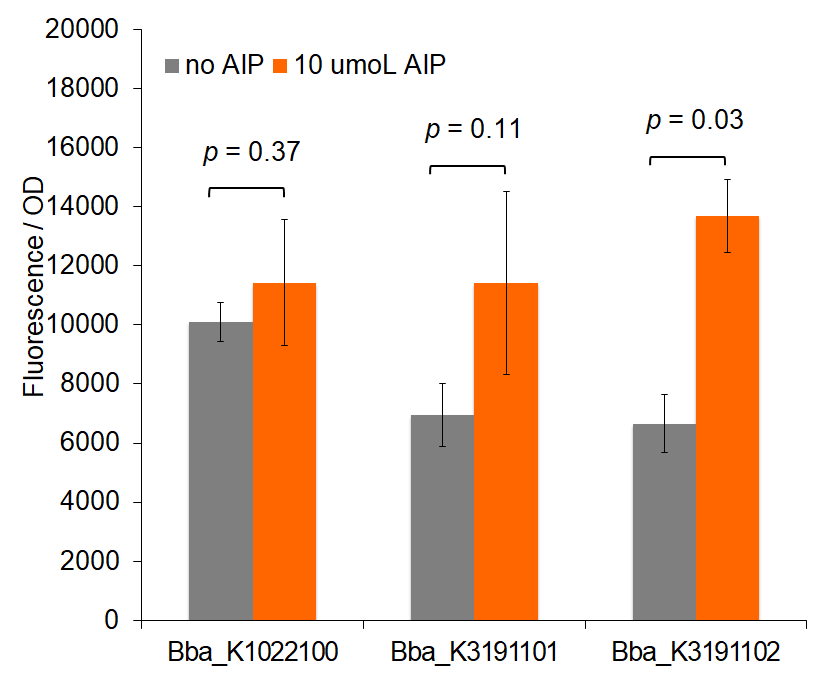
Figure 3: Total change in fluorescence of permeabilized cells with and without AIP induction.
Results
Our characterization data displays the difference in fluorescence values between BBa_K206000, BBa_K1022100, BBa_K3191101, and BBa_K3191102. This data was analyzed with a two-tail t-test of unequal variances with a significance level of 0.05 to draw conclusions of significance. Based on these values, fluorescence for BBa_K1022100 and BBa_K3191101 tend to exceed the control. This demonstrates that BBa_K1022100 and BBa_K3191101 both function properly.
Figure 1 demonstrates fluorescence values under the pre-permeabilization protocol. These data indicate that AIP induction caused minimal difference in fluorescence intensity. Figures 2 and 3 show fluorescence intensity and change in fluorescence, respectively, of cells after the permeabilization protocol. When comparing total fluorescence, BBa_K1022100 tends to yield significantly greater fluorescence values than BBa_K3191101 at hour 8, as supported by a p-values of 0.002 for the AIP trial. This suggests the modified sequences, while more complete, do not successfully increase GFP production under the original characterization procedures. The addition of SarA also does not appear to provide a significantly greater fluorescence under the influence of AIP. This is seen in a comparison of BBa_K3191101 and BBa_K3191102 at hour 3 in figure 4 (p-value = 0.902).
Trends in the data suggest that the introduction of AIP appears to have a greater effect on BBa_K3191101 and BBa_K3191102 than on BBa_K1022100 under the permeabilization characterization conditions. The fluorescence values for these conditions are seen in figures 2 and 3. In these conditions, a Tris-EDTA solution is used to attempt to increase AIP intake by the cells. BBa_K1022100 does not experience significantly greater fluorescence than the control at hour 3 with AIP induction (p-value = 0.111). In comparison, BBa_K3191101 and BBa_K3191102 have significantly greater fluorescence at hour 3 with AIP induction. This is demonstrated with p-values of 0.010 for BBa_K3191101 and 0.002 for BBa_K3191102. This suggests that AIP induction does not significantly increase fluorescence in BBa_K1022100, while it does increase in BBa_K3191101 and BBa_K3191102.
The increase in fluorescence may be explained by the modified experimental procedure. If the presence of Tris-EDTA successfully increases AIP uptake, then an increase in AIP uptake only improves fluorescence in BBa_K3191101 and BBa_K3191102. This suggests that our parts are truly induced by AIP, while BBa_K1022100 is not.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 125
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 569
Illegal BamHI site found at 65 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1848
Illegal BsaI.rc site found at 1896
Illegal BsaI.rc site found at 3159
| None |
