Part:BBa_K2934006
AmyE - Invertase-Histag for Bacillus subtilis
This part comprises of the Invertase gene (BBa_K2934001) connected to the secretory signal peptide AmyE (from the alpha-amylase gene) (BBa_K2273023), using a short nine amino-acids linker BBa_K2934005). This part was constructed in our lab using the commercial plasmid pBE-S (TaKaRa) as a backbone.
Unfortunately, the enzyme Invertase was not secreted and active with this specific signal peptide. Therefore, we performed the same experiment with a different plasmid, that contained the same Invertase gene but with a library of 174 signal peptides instead, and in this experiment, we were able to detect extracellular enzymatic activity. For full description, see the Invertase part page (BBa_K2934001) and our lab results.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 989
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1294
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 595
Illegal BsaI.rc site found at 937
Illegal SapI.rc site found at 1197
Characterization
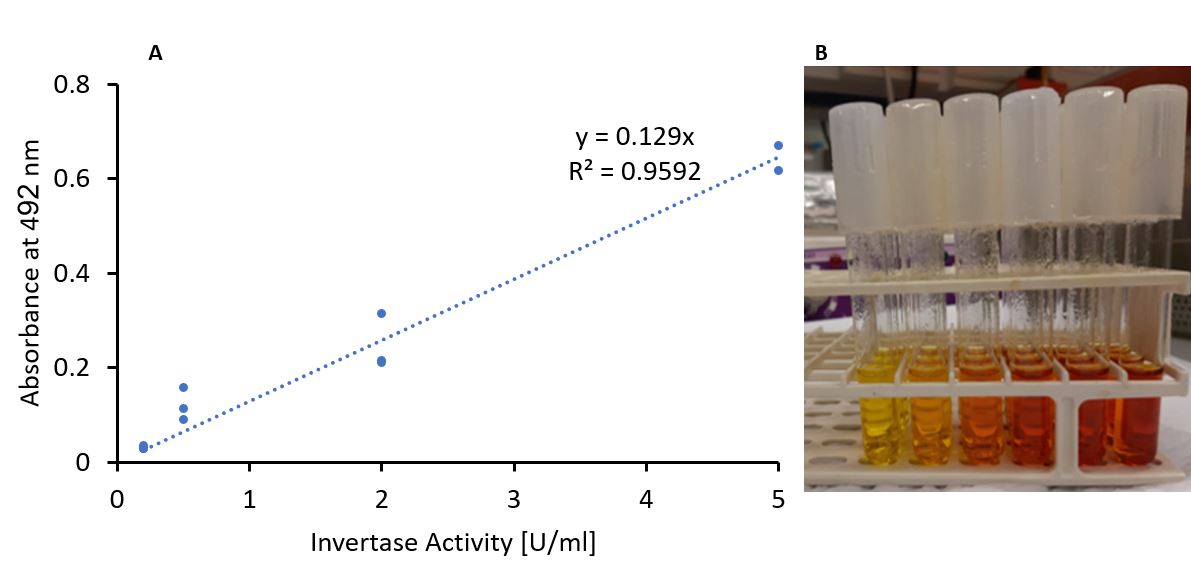
Invertase is an enzyme that cleaves sucrose to form glucose and fructose. Thus, a precise method that will detect either glucose or fructose can indicate the amount of sucrose cleaved by the enzyme. The test we used is based on the "reducing sugars" assay, in which reducing sugars react with the yellow picric acid to form picramic acid, which is red (Figure 2). Therefore, the absorbance of red light (492nm) could indicate the amount of sucrose cleaved, which indicates the enzymatic activity.
To verify that the activity-absorbance correlation remains linear under the experiment is conditions, we performed the experiment using a commercial enzyme, creating a calibration curve. We used increasing concentrations of commercial invertase (with a well-determined specific activity) and a constant saturated concentration of sucrose. Then, we checked the 492 nm light absorbance and plotted the calibration curve (Figure 3).
After the cloning process had been completed, we performed an activity test on the bacterial invertase. We tested both bacterial lysate and bacterial supernatant to determine whether active invertase had been produced and secreted by evaluating the activity in each sample. The recombinant bacteria samples were compared to wild type (WT) samples.
The first colony tested contained the invertase enzyme with AmyE as a signal peptide. Unfortunately, no notable activity was detected (Figure 4).
These results, in addition to the results observed in the SDS-PAGE experiment, indicating that the AmyE signal peptide may interfere with either the enzyme's production or its activity.
Methods
This vector was constructed by Gibson assembly, with the pBE-S commercial plasmid (Takara) that served as a backbone, and eventually transformed into Bacillus subtilis (RIK1285).
The bacteria were cultured over-night at 37°C in a shaking incubator (250 rpm), followed by 1:100 dilution. The diluted culture was incubated for another few hours while monitoring the culture turbidity by measuring the O.D600.
When the culture has reached the logarithmic growth phase, the bacterial extracellular and intracellular proteins were separated, after collecting the supernatant and the pellet (followed by lysis, using sonicator).
For further reading of our full protocols (cloning, enzymatic assays and enzymatic assay under different pH levels) look at our wiki protocols page.
| None |