Part:BBa_K2239007
CBD-7betaHSDH
CBD--7beta-HSDH (T7 promoter--lac operator--RBS--His-tag--7beta-HSDH--CBD--T7 terminator)
This device codes for the 7beta-HSDH--CBD fusion protein.
Construct
The vector of 7beta-HSDH--CBD for its expression is pET-28x. It is formed by modifying the restriction enzyme sites EcoR I and Xba I of vector pET-28a.
The 7beta-HSDH sequence is retrieved from the GenBank. It is artificially synthesized and inserted into plasmid pUC57. The 7beta-HSDH gene is then cloned from the plasmid by PCR amplification. The restriction site BamH I is added to the upstream primer, and Hind III is added to the downstream primer.
The CBD sequence is retrieved from the GenBank. It is artificially synthesized and inserted into plasmid pUC57. The CBD gene is then cloned from the plasmid by PCR amplification, with the restriction site Hind III added to the upstream primer, and Xhol I added to the downstream primer.
Firstly, the 7beta-HSDH gene is inserted into the modified pET-28x at BamH I and Hind III, and CBD at Hind III and Xhol I, after proliferation in T3 vector. Then the whole gene fragment, T7 promoter--lac operator--RBS--His-tag--7beta-HSDH--CBD--T7 terminator, is retrieved from this plasmid by PCR amplification, with prefix containing EcoR I, Not I and Xba I added on its upstream primer, and suffix containing Pst I, Not I and Spe I added on its downstream primer. The PCR product is then connected to pSB1C3 at EcoR I and Pst I.
[Fig. 1. pSB1C3--CBD--7beta-HSDH]
Usage and Biology
7beta-HSDH(7beta-hydroxysteroid dehydrogenase) catalyzes the reduction of hydroxysteroids at C-7 position and back, as it converts NADPH to NDAP+ and back. It catalyzes the reduction of the intermediate product 7-oxo-LAC (7-ketolithocholic acid) into UDCA, the final product, where the carbonyl at C-7 position becomes hydroxyl.[1]
CBD (cellulose binding domain) is able to bind to cellulose. When connected to 7beta-HSDH, CBD is able to immobilize the enzyme 7beta-HSDH after expression, by binding to the gauze inside the solution on its cellulose.[2]
The function of cellulose binding domain
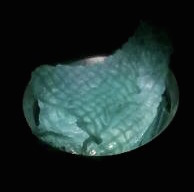
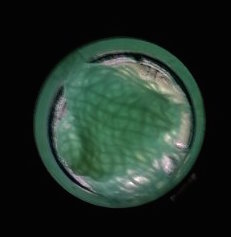
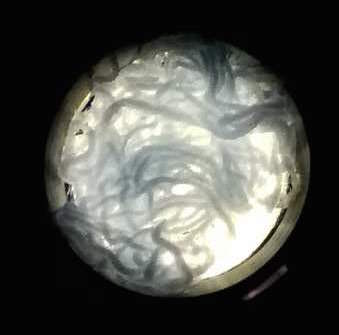
The function of CBD is tested by connecting CBD gene with GFP gene in pET28x. The GFP-CBD fusion protein is expressed and mixed with a gauze piece. The green fluorescent on the gauze is not significantly reduced after washing, proving that the CDB is well functioned. In comparison, no green fluorescent is left after washing the gauze mixed with GFP-ChBD (Chintin binding domain).
[Fig. 2. GFP-CBD on gauze before washing]
[Fig. 3. GFP-CBD on gauze after washing]
[Fig. 4. GFP-ChBD on gauze before washing]
[Fig. 5. GFP-ChBD on gauze after washing]
Expression and Immobilization[2]
The constructed pET28x--7beta-HSDH--CBD plasmid is transformed into BL21(DE3) E.coli for expression. After that, when the OD 600 reached 0.6-0.8, 0.2mM IPTG is added in the liquid culture. The mixture is shaken at 20 ℃ overnight. The bacteria is collected by centrifugation at low temperature, 8000 rpm for 10 minutes, and the supernatant is discarded. The bacteria is then resuspended using 0.15M pH8.8 Tris-HCL, and is broken by ultrasonication.
The resulted bacteria solution is diluted to a certain concentration and mixed with gauze piece, and the gauze piece is washed three times by ddH2O afterwards. As a result, the CBD protein binds to the cellulose on gauze, and the enzyme is successfully immobilized.
Enzyme Activity
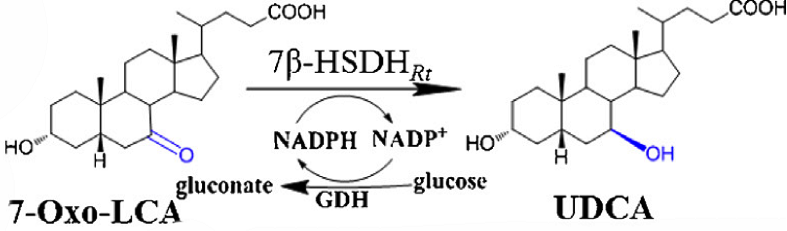
The reduction of 7OXO-LCA to UDCA
[Fig. 6. Reaction process]
7β-HSDH-CBD is an NADPH dependent enzyme from Ruminococcus Torques. The 3mL reaction contains 150mM phosphate buffer(pH 8.0), 10mM UDCA, 0.2mM NADPH. The reaction started when the solution is combined with 7β-HSDH-CBD-enzyme-binding gauze that in different concentration, includes 80ul, 120ul, 160ul liquid supernatant of ultrasonication bacteria solution. The control group was testify under the same solution and condition but using pET28x-CBD liquid supernatant of ultrasonication bacteria solution to bind with gauze in the concentration of 160ul. Before adding the gauze into the solution, the gauze was washed by ddH2O for 3 times in order to purify the enzyme.
The 7-OXO-LCA was convert to UDCA by Taking a a pair of hydrogen(2H+and 2e-)from the NADPH and form 7-hydroxyl group (B position). The co-enzyme NADPH is the donor of the a pair of hydrogen(2H+and 2e-), and was transformed into NADP+. However, to make the reaction easy to testify, the reverse reaction that transforming UDCA to 7OXO-LCA was employed to prove the function of 7B in which The B position 7-hydroxyl group loose a pair of hydrogen(2H+and 2e-) and from the 7-carbonyl group, and NADP+ is the acceptor of the a pair of hydrogen(2H+and 2e-), to form NADPH The the synthesized NADHP can be determined spectrophotometrically at340 nm (ε = 6.22 mM-1 cm-1) and room temperature. One unit of activity is defined as the amount of enzyme catalyzing the synthesize of 1 mmol of NADPH per min under the assay conditions used.
Result
[Fig. 7. Result]
Reduction of 7-oxo-LCA to UDCA using 7β-HSDH and GDH(NADPH regeneration)
[Fig. 8. Reaction process]
The 3mL reaction solution containing 150 mM phosphate buffer(pH 8.0), 10 mM UDCA, 30 mM glucose, 0.2mM NADP+, combined with 1U/ml 7β-HSDH and 5U/ml GDH at room temperature.
The bioconversion experiment was monitored via HPLC measurements. The sample was analyzed by UV detection at 210nm. We testify the synthesize of 7-oxo-LCA and the decrease of UDCA, using a mobile phase of methanol–water mixture (final ratio 80:20,pH 3.5 with phosphoric acid) using chromatographic column C18 .
Result
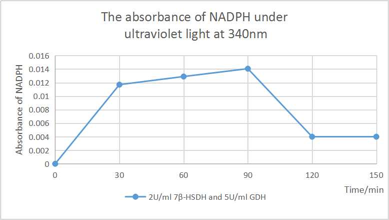
[Fig. 9. Absorbance of NADPH]
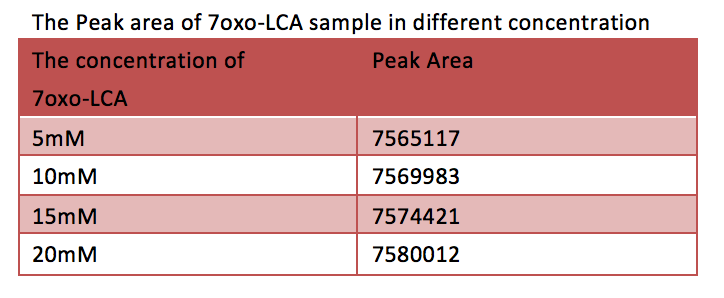
[Fig. 10. Peak area of 7oxo-LCA sample in different concentration]
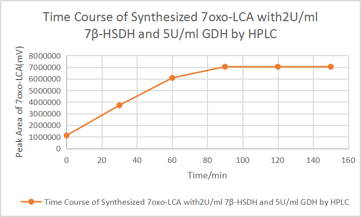
[Fig. 11. Time course]
According to the HPLC result after 90minute, there is no significant increase of 7oxo-LCA, as a result, most of the UDCA has been transformed into 7oxo-LCA. And according to the HPLC the final yielding rate is 93%.
According to the Absorbance of NADPH that shown in figure, the absorbance is decreased significantly after 90 minute dual to the depleted UDCA that stop the conversion of UDCA to 7oxo-LCA and NADP+ synthesize. Because of the abundant amount of glucose in the solution, the GDH that works on glucose still regenerate the NADPH by taking a pair of hydrogen from glucose until most of the NADP+ transformed into NADPH.
Reference
[1] Ming-Min Zheng, Ru-Feng Wang, Chun-Xiu Li, Jian-He Xu: Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β-hydroxysteroid dehydrogenase from Ruminococcus torques. Process Biochemistry, Elsevier, 2015.
[2] Etai Shpigel, Arie Goldlust, Gilat Efroni, Amos Avraham, Adi Eshel, Mara Dekel, Oded Shoseyov: Immobilization of Recombinant Heparinase I Fused to Cellulose-Binding Domain, 1999.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1122
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 537
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 978
| None |