Part:BBa_K1483001
Endo-β-Galactosidase
Encodes for an enzyme, that is capable of cleaving A- and B-antigens in the human ABO-Bloodgroup system. As the name suggests, the galactose and connected saccharide residues are cleaved from the antigen, leaving behind the the Oh-Antigen which is characteristic of the Bombay-blood type. Part in RFC25.
Contributions by 2021 TAS_Taipei
We obtained the amino acid sequence of the Endo-β-Galactosidase protein, derived from Clostridium perfringens, from the iGEM DNA Repository Plate (BBa_K1483001), as entered into the iGEM parts collection database by the Tuebingen iGEM team in 2014.
In order to test protein expression of the enzyme, we added a strong promoter and strong ribosome binding site (RBS; BBa_K880005) upstream of the protein amino acid sequence to create a part BBa_K3717005.

Figure 1. Colony PCR check for strong promoter (K88) Endo-β-Galactosidase (Part:BBa_K3717005) using VF2 and VR primers. Confirms successful ligation as a band is produced at the expected size of 2752bp, as indicated by the red triangle.
We tested protein expression of the composite part BBa_K3717005 by transforming our plasmids into BL21 E. coli cells. We grew cultures at 37°C overnight, diluted those cultures, and then grew to OD600 0.5~0.6 at 37°C. We then induced expression with 0.5 mM IPTG and allowed cultures to grow overnight at room temperature. We took samples pre-induction and post-induction and examined them by SDS-PAGE.
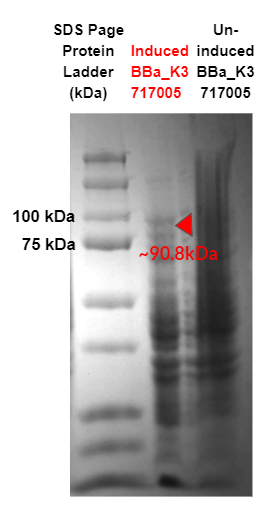
Figure 2. SDS Page of cell lysate for the strain: strong promoter (K88) Endo-β-Galactosidase (Endo-β-gal) (Part:BBa_K3717005). Red triangles indicate expected size for Endo-β-gal (90.8 kDa). Sequences for target proteins were found to not contain a start codon and thus have no expression, as shown by the triangles.
Our SDS-page in Fig. 2 did not show any overexpression bands for the enzymes of interest. The results indicate that there were no target proteins at their expected band sizes: 90.8 kDa band for K88 promoter + Endo-β-gal in the induced sample. As the SDS page is of cell lysis samples, other bands present are due to innate proteins present in the bacteria cell.
Upon comparison of the amino acid sequence from Tuebingen’s parts with expected full sequences that were obtained from UniProt, we discovered that the enzyme sequences were missing the start codon (Fig. 3), which explained the non-expression of the proteins.
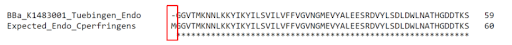
Figure 3. Top sequence: First 59 amino acids of Team Tuebingen's 2014 Endo-β-gal part BBa_K1483001. Bottom sequence: First 60 amino acids of expected sequence for Endo-β-gal derived from Clostridium perfringens. Based on the alignment of the two sequences, Tuebingen's part is missing the first amino acid (start codon) of the Endo-β-gal protein.
We improved their part by adding the missing base sequences, including the start codon, and attached a 6x Histidine tag through a flexible Glycine-Serine linker upstream of the protein sequence (Fig. 3). We derived the sequence of Endo-β-Galactosidase from Streptococcus pneumoniae that was optimized to increase the activity of the enzyme by around 170-fold [1]. This formed our new Endo-β-Galactosidase basic part (Part:BBa_K3717008).

Figure 4. Open reading frame for Endo-β-Galactosidase with N-Terminal 6x Histidine tag (BBa_K3717008)
We used this to create a new composite part (Part:BBa_K3717011) through the attachment of a T7 promoter and strong RBS (BBa_K525998) upstream of the open reading frame, as well as a double terminator (BBa_B0015) downstream of the ORF.
 )
)
Figure 5. Design of Endo-β-Galactosidase with T7 Promoter, strong RBS, N-Terminal 6x Histidine tag and Double Terminator Construct (Part:BBa_K3717011)
References
1. Kwan, David H., et al. “Toward Efficient Enzymes for the Generation of Universal Blood through Structure-Guided Directed Evolution.” Journal of the American Chemical Society, vol. 137, no. 17, American Chemical Society, May 2015, pp. 5695–705. ACS Publications, https://doi.org/10.1021/ja5116088.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
