Part:BBa_K814002
dehydroquinate O-methyltrasferase (O-MT) generator
Our research has focused on two novel biosynthetic pathways found in two distinct algal species. A pathway ending in the production of two UV-protective compounds, shinorine and mycosporine-glycine, was cloned from Anabaena varibalis. Dehydroquinate O-methyltransferase (O-MT) catalyzes the second step in this pathway, converting dehydroquinate to 4-deoxygadusol.
This part includes a modified constitutive lac promoter (lacPmod), and RBS and the open reading frame of O-MT. This part can be used to express O-MT in E. coli.
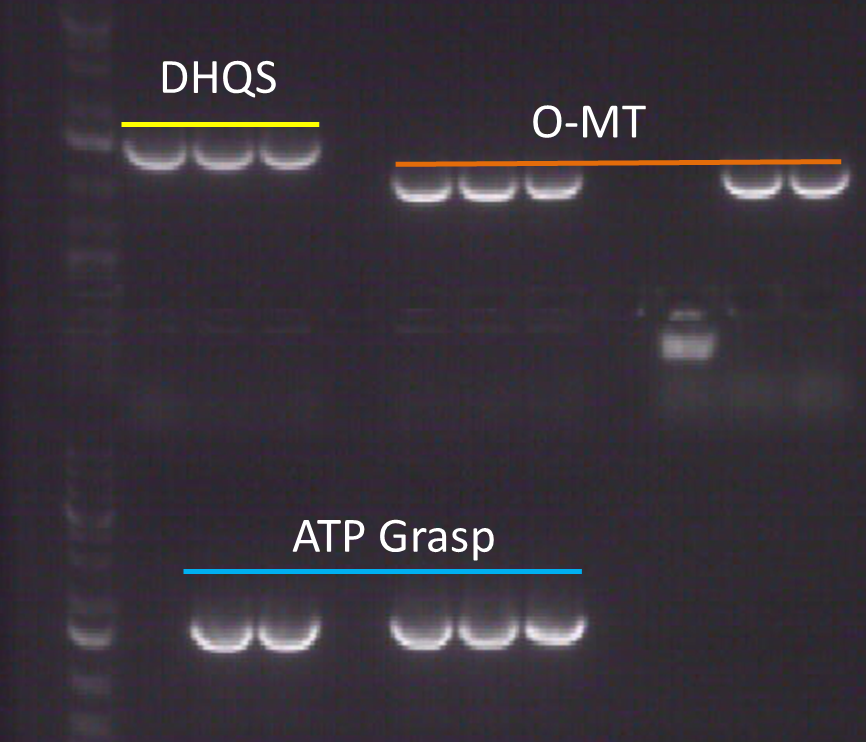
Figure 1. PCR screening of O-MT from colonies following ligation and transformation into the pUCBB plasmid.
Balskus and Walsh, 2010. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3116657/]
BBa_K3634001
Codon optimised O-methyltransferase (O-MT) CDS for use in E.coli as part of the shinorine synthesis gene cluster. By using the IDT codon optimisation tool, St Andrews iGEM 2020 have optimised the GC content of the sequence taken from A.variabilis to improve production efficiency of the final product shinorine. We have further made the part biobrick assembly standard RFC[10] & RFC[1000] compatible by removing XbaI and SapI restriction sites introduced by this optimisation step. The part should be used alongside the additional optimised parts (BBa_K3634000, BBa_K3634002 and BBa_K3634003) responsible for the ultimate conversion of the substrate sedoheptulose 7-phosphate to the final product shinorine.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 418
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 980
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 134
Illegal XhoI site found at 988 - 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 418
- 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 418
Illegal AgeI site found at 518 - 1000COMPATIBLE WITH RFC[1000]
| None |