Part:BBa_K786001:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Characterization of BBa_K786001
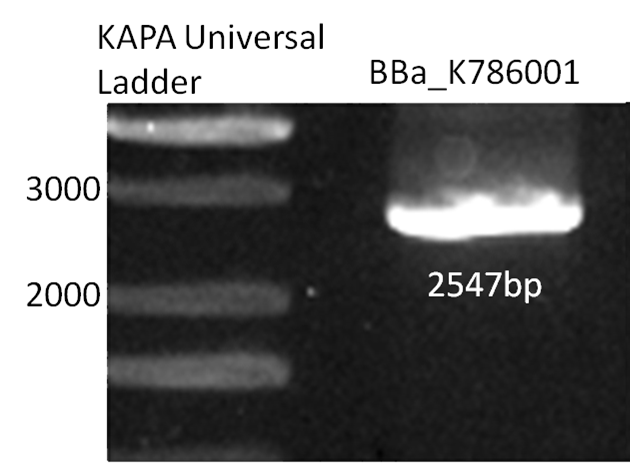
1. Biobrick DNA length verification
In order to check the inserted biobrick DNA length, PCR was performed with standard biobrick vector sequencing primers (BBa_G00100 and BBa_G00101).
The PCR product meets the target size (2547bp), the DNA sequence was further verified by DNA sequencing.
2. Promoter efficiency for BBa_K786001. BBa_K786002, BBa_K786003
To test the expression of sensory rhodopsin triggered by constitutive promoter BBa_J23100 to sense light,
we need to test the effect conferred by different E. coli strains to expression of red fluorescence protein reporter downstream of BBa_J23100 to different bacterial strains. It allows us to select the suitable strain(s) for this constitutive promoter for expressing sensory rhodopsin.
Florescence plate reader was used to take readings of fluorescence emission of 635nm and absorbance at 600nm (OD600) between time intervals of 12 hours on each strain. The measurements were started when the cultures reached a OD600 of around 0.4 that represents log phase of active proliferation. Growth curve and fluorescence intensity against time were plotted to compare cell growth and protein expression on different strains.
Three independent experiments were conducted. No significant difference was observed on the growth curves, indicating a similar growth rate among the three bacterial strains with BBa_J23100 transformed. It implies the promoter does not cause cell toxicity or growth inhibition of these three bacterial strains.


For the protein expression, the results showed that the fluorescence intensity of reporter in DH5α was significantly lower compared with TOP10 and BL21(DE3).
To conclude, DH5α is not an optimal strain to utilize promoter BBa_J23100, while TOP10 and BL 21(DE3) can effectively express the reporter. Therefore, in downstream application of our light sensing biobricks (BBa_K786001, BBa_K786002, BBa_K786003) in which BBa_J23100 was used, DH5α are not used.
3. Protein expression verification
A poly-histidine DNA sequence with a linker(GGGAAGCTT-atgcatcatcatcatcatcat-GGTGGTGGT- AAGCTTGGG)was ligated before the SRII gene sequence by using the HindIII site. Then the construct was transformed into different strains of E.coli.(BL21, TOP10, Rosseta 2 and DH5α.
Protein expression of each strian was tested by western blotting analysis using anti-his antibodies.
BBa_K786001 was expressed in Rosetta 2, BL21 (DE3) and TOP10 only, but not in DH5α.
Future Application
Together with Positive Phototactic Construct for Blue Light Detection (BBa_K786002), Phototactic Construct for Orange Light Detection BBa_K786003, Light-triggered Gene Expression System BBa_K786010 and the genome targeting CRISPR/Cas systems BBa_K786031. This part will have the following applications.
I. Light-directed bacterial cells sorter
When the phototactic systems are integrated with heavy metals collection devices [1, 2] , such as SmtA and MntH for collecting cadmium ions, fMT and Glpf for collecting arsenic ions, and BBa_K346005, a mercury(II) ions absorption device, our sensory rhodopsin systems could become a useful tool for heavy metal sorting in present in sewage.

When different ion-absorption systems are integrated with our Sensory Rhodopsin systems, different kinds of ions can be directed to different position for heavy metals sorting and separation in sewage, which could be a new tool for heavy metal recycling.
Metal ions collection aided by light-directed cell sorting in an environmental friendly way
Other than integrating with heavy metal absorption device, our sensory rhodopsin system can also be incorporated with our gene expression system and our genome targeting system in order to sort out and collect heavy metal ions while minimizing potential environmental hazards due to release of genetically engineered organisms.

First of all, the cells absorbing two different ions can be sorted out by blue light alone with positive phototactic device (BBa_K786002) and negative phototactic device (BBa_K786001).
An orange light source can then trigger the genome targeting system (BBa_K786031) together with the gene expression device for sensory rhodopsin (BBa_K786010).
The cells will eventually destroy their own genetically engineered DNA, and leading to cell death and releasing the heavy metal ions. Furthermore, this step can prevent horizontal gene transfer due to the disposal of cell debris to the environment, minimizing the environmental impact.
II. Improvement on the HR desalination system
Our sensory rhodopsins system (BBa_K786003) and the halorhodopsin system (BBa_K559010) are both sensitive to long wavelength visible light source. They can be integrated to enhance the desalination efficiency [3]. As E. coli can be attracted to a region closer to the light source with greater intensity, the halorhodopsin system should function with a higher efficiency and absorb more Cl- ions.
BBa_K786002 the phototactic device sensitive to near-UV light can be integrated to become a tool for cells/Cl- ions direction to a separate chamber, so that the remaining solution can achieve desalination.
III. Incorporation with BBa_K786031 as a new biosafety approach
Genetically-modified organisms have been put in use extensively throughout the last decade. Yet, concerns about such organisms causing undesirable effect to the environment once released have been raised. Our biobricks might be one of the answers to tackle such concerns. When the gene expression system for sensory rhodopsin is incorporated with the CRISPR/Cas system, bio-engineered cells would cleave their own DNA into nucleotides once exposed to natural light.
This system would be useful for bio-engineered cells that could be used to work in the dark. (e.g.: Biofuel synthesizing cells, human hormone-producing cells)
[1] PKU 2010 iGEM Team
[2] Tokyo-NoKoGen 2010 iGEM Team
[3] CUHK 2010 iGEM Team
User Reviews
UNIQ72d940717e6b6c0b-partinfo-00000000-QINU UNIQ72d940717e6b6c0b-partinfo-00000001-QINU